Deposition Date
2012-03-30
Release Date
2012-07-11
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4AOV
Keywords:
Title:
DpIDH-NADP. The complex structures of Isocitrate dehydrogenase from Clostridium thermocellum and Desulfotalea psychrophila, support a new active site locking mechanism
Biological Source:
Source Organism(s):
DESULFOTALEA PSYCHROPHILA (Taxon ID: 84980)
Expression System(s):
Method Details:
Experimental Method:
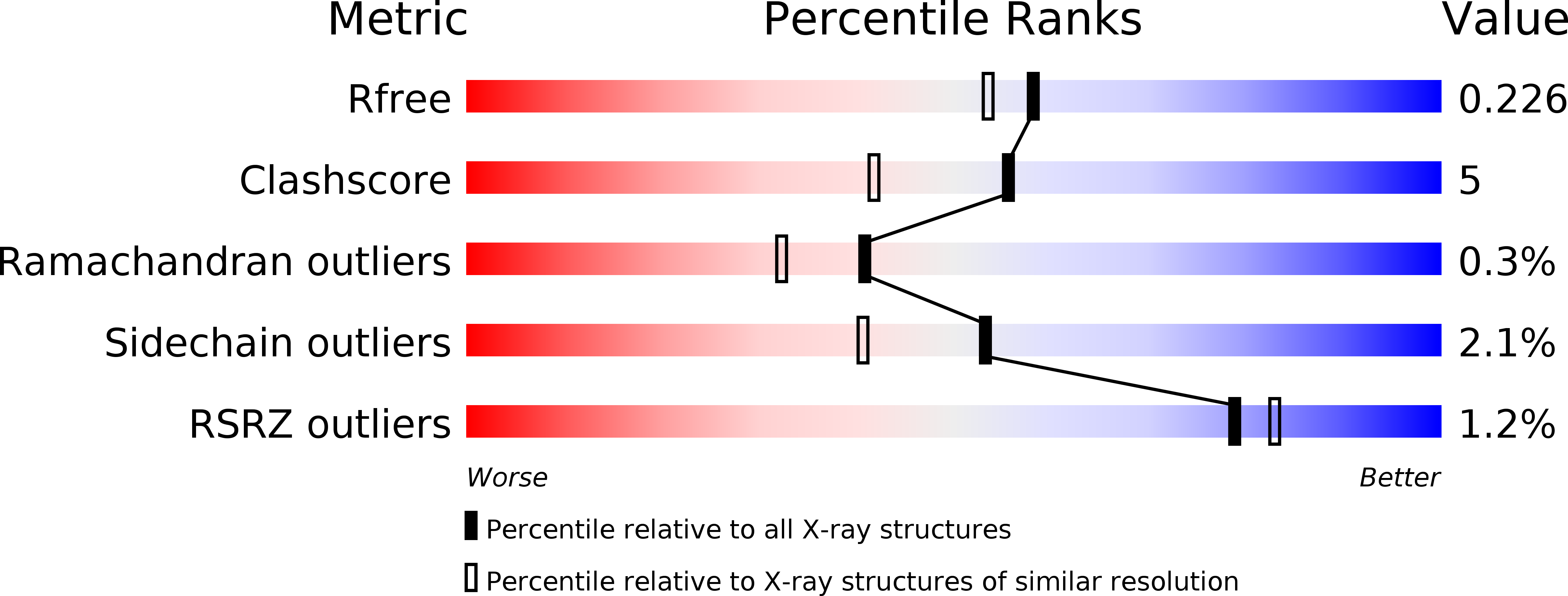
Resolution:
1.93 Å
R-Value Free:
0.22
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
C 1 2 1