Deposition Date
2012-03-15
Release Date
2012-08-29
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4AN5
Keywords:
Title:
Capsid structure and its Stability at the Late Stages of Bacteriophage SPP1 Assembly
Biological Source:
Source Organism(s):
BACILLUS PHAGE SPP1 (Taxon ID: 10724)
Expression System(s):
Method Details:
Experimental Method:
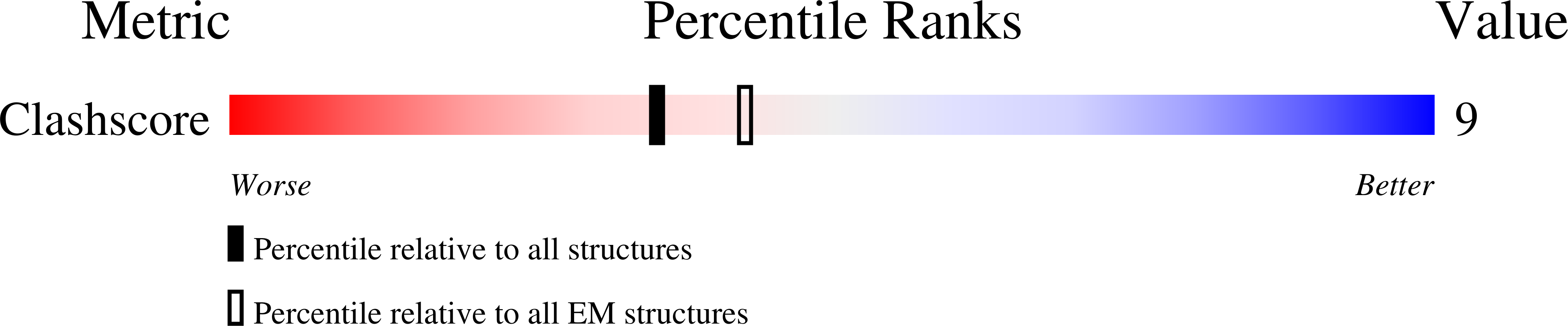
Resolution:
8.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE