Deposition Date
2012-02-06
Release Date
2013-02-20
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4AHA
Keywords:
Title:
Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with fucosylated monosaccharides (Fuc1-2Gal, Fuc1- 3GlcNAc, Fuc1-4GlcNAc and Fuc1-6GlcNAc)
Biological Source:
Source Organism(s):
ASPERGILLUS FUMIGATUS (Taxon ID: 5085)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.20 Å
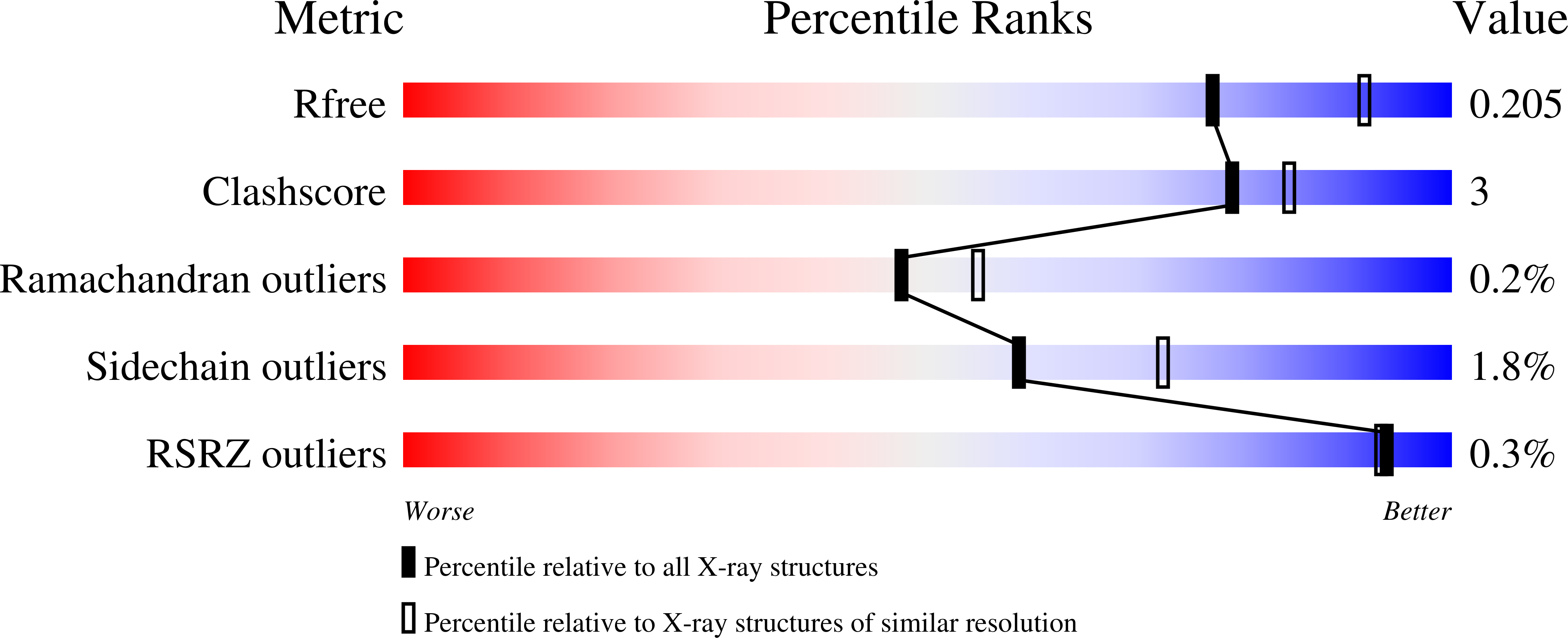
R-Value Free:
0.21
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 1 21 1