Deposition Date
2011-12-01
Release Date
2012-05-02
Last Version Date
2023-12-20
Entry Detail
PDB ID:
4AAD
Keywords:
Title:
Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in absence of metal ions at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3')
Biological Source:
Source Organism(s):
CHLAMYDOMONAS REINHARDTII (Taxon ID: 3055)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
SYNTHETIC CONSTRUCT (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
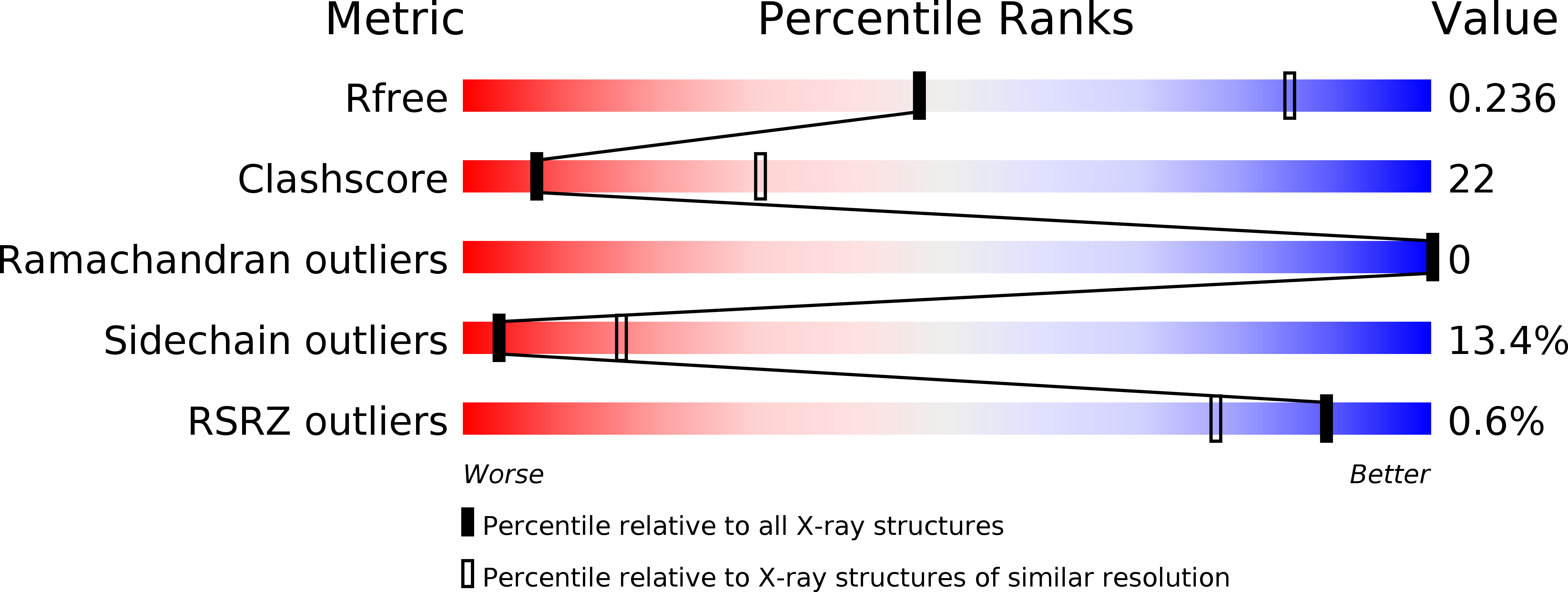
Resolution:
3.10 Å
R-Value Free:
0.23
R-Value Work:
0.20
R-Value Observed:
0.21
Space Group:
P 2 21 21