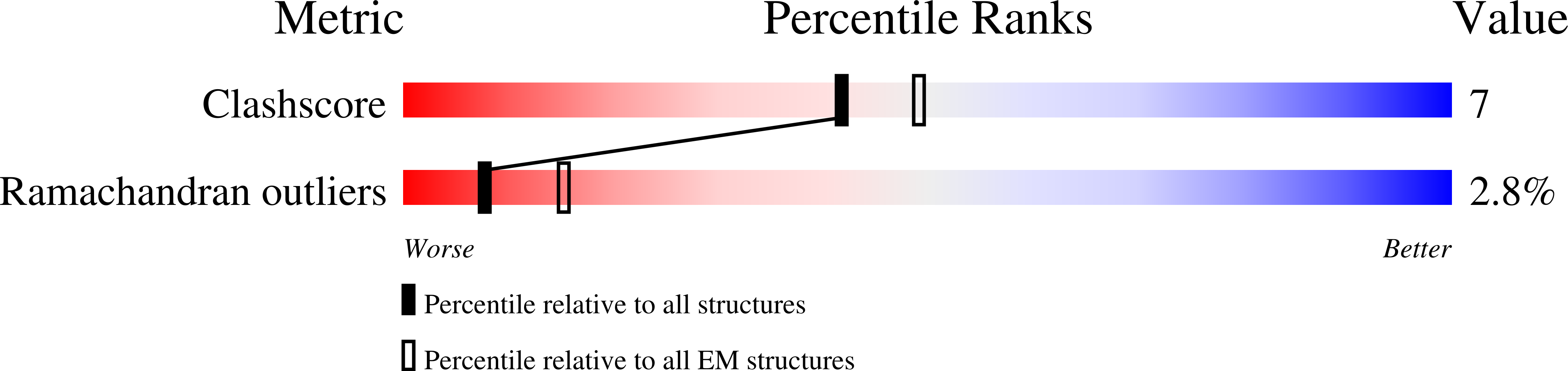Deposition Date
2011-11-18
Release Date
2012-01-11
Last Version Date
2024-05-08
Entry Detail
PDB ID:
4A82
Keywords:
Title:
Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966.
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
Method Details:
Experimental Method:
Resolution:
9.00 Å
R-Value Work:
0.37
Space Group:
P 1