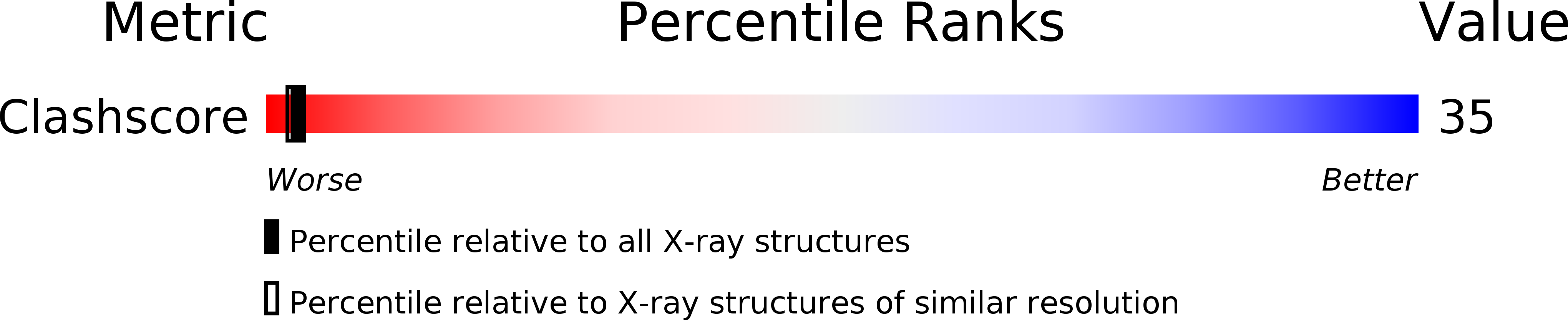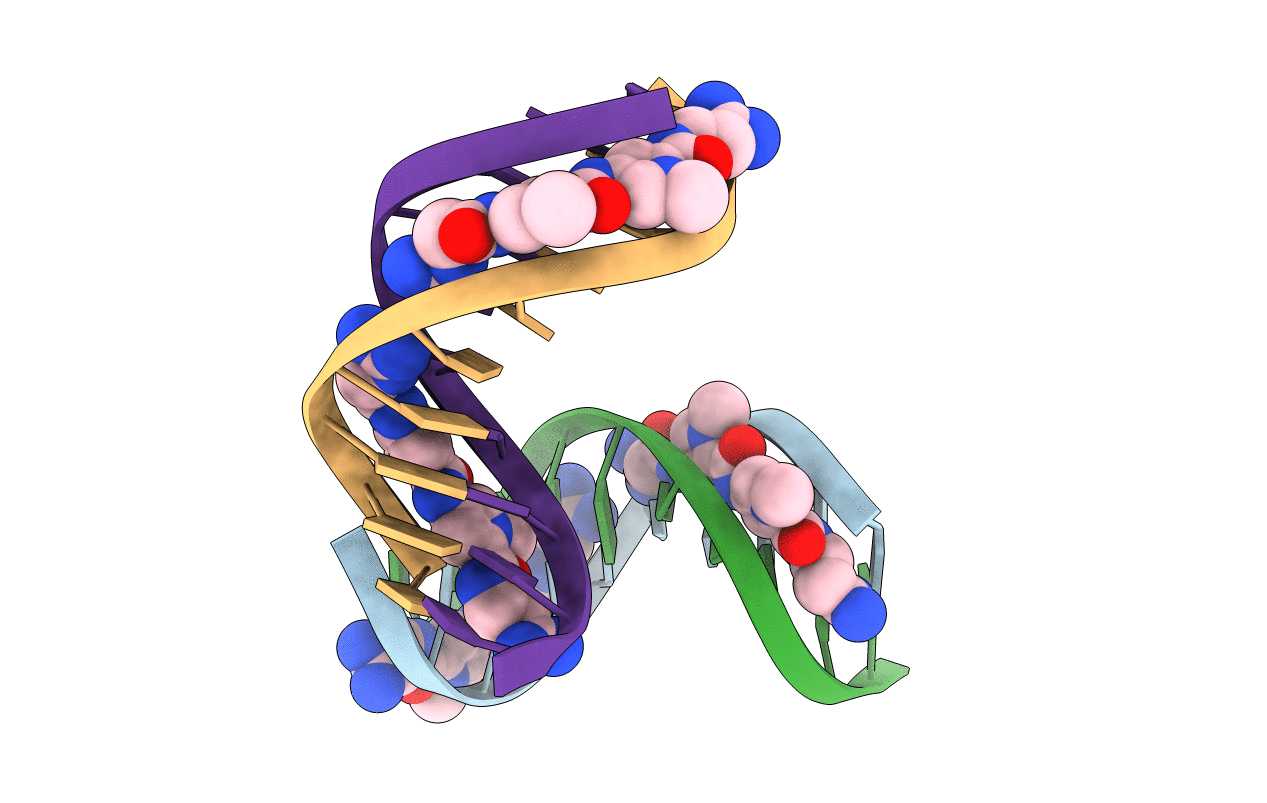
Deposition Date
1998-01-14
Release Date
1998-12-02
Last Version Date
2024-02-28
Entry Detail
PDB ID:
474D
Keywords:
Title:
A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2
Method Details:
Experimental Method:
Resolution:
2.40 Å
R-Value Free:
0.27
R-Value Work:
0.19
Space Group:
P 1