Deposition Date
1999-01-05
Release Date
2001-09-14
Last Version Date
2023-12-27
Entry Detail
PDB ID:
439D
Keywords:
Title:
5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'
Method Details:
Experimental Method:
Resolution:
1.60 Å
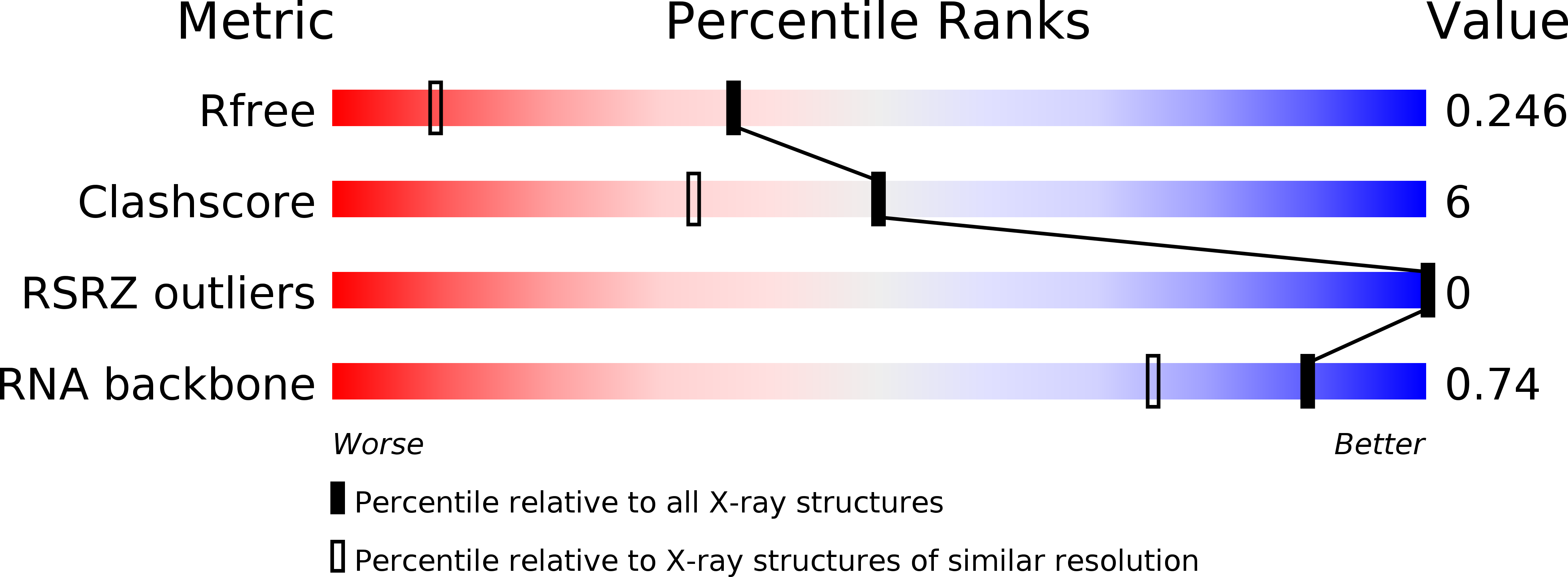
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
H 3 2