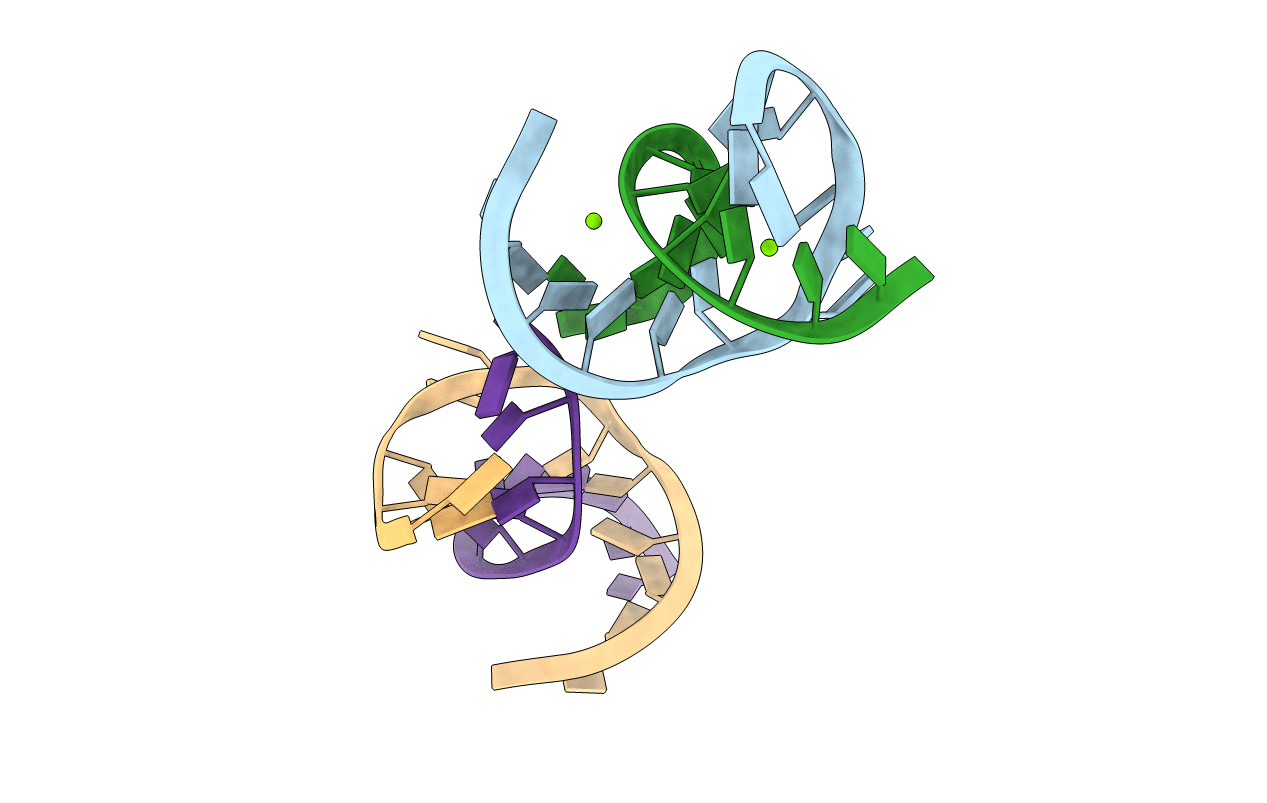
Deposition Date
1998-09-29
Release Date
1999-03-01
Last Version Date
2024-02-28
Entry Detail
PDB ID:
429D
Keywords:
Title:
CRYSTAL STRUCTURE OF A LEADZYME; METAL BINDING AND IMPLICATIONS FOR CATALYSIS
Method Details:
Experimental Method:
Resolution:
2.70 Å
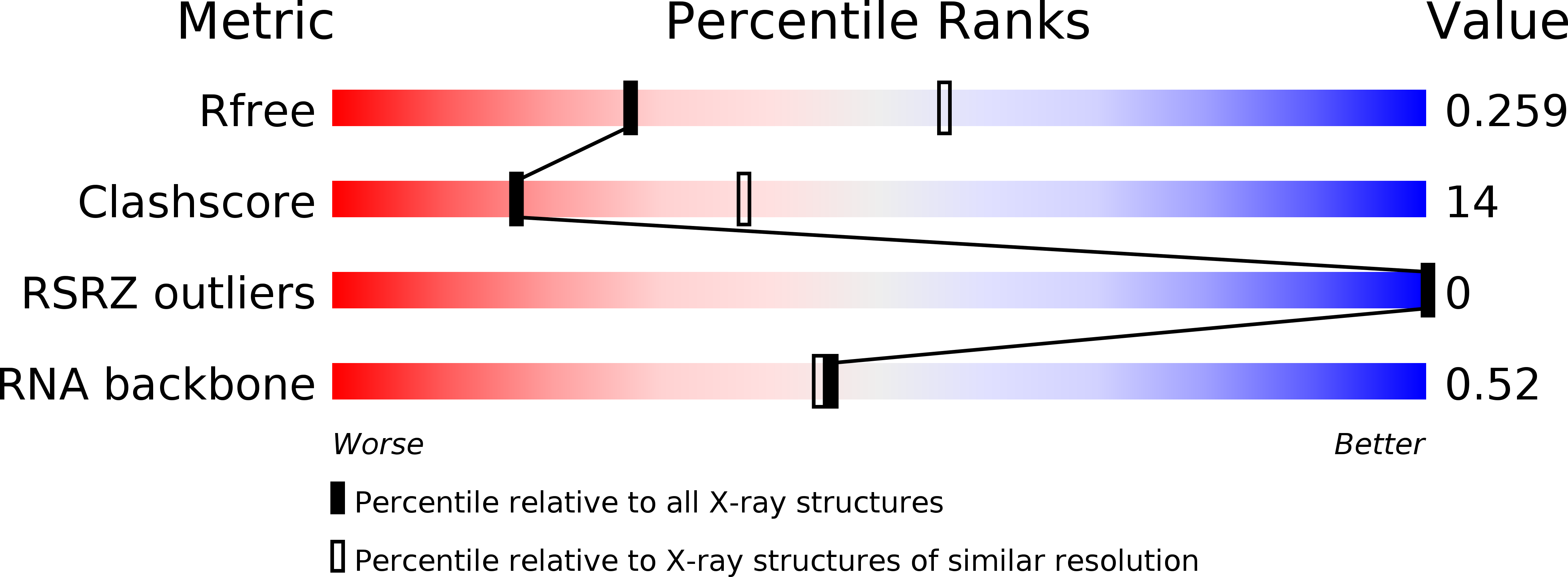
R-Value Free:
0.27
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
P 61 2 2