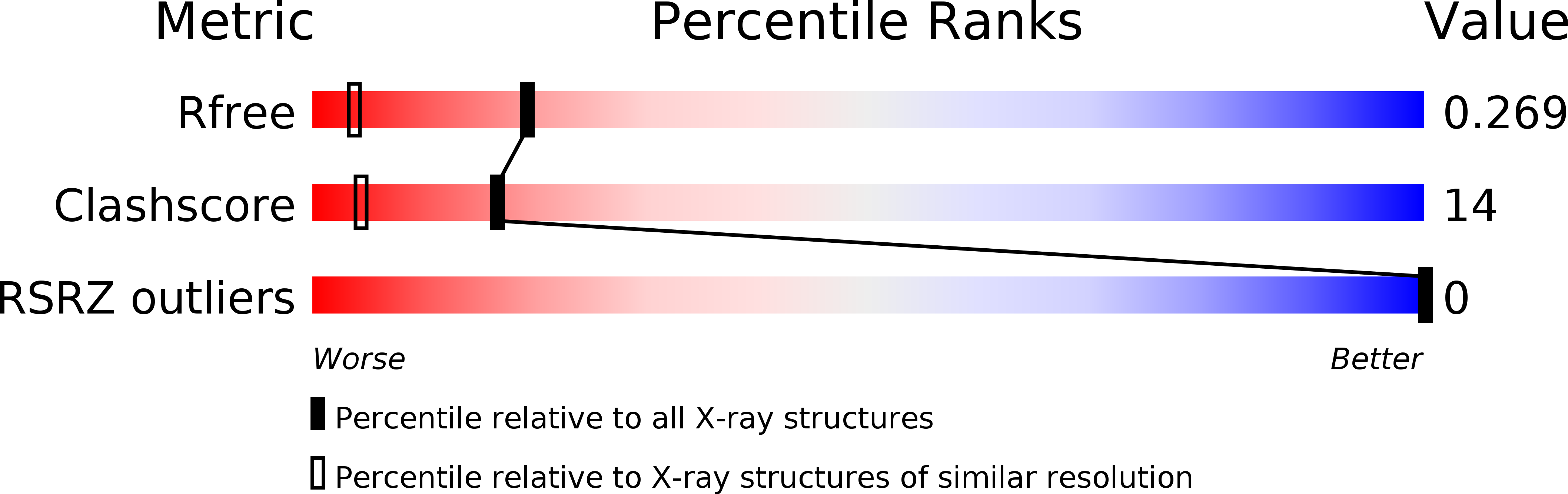Deposition Date
1998-09-28
Release Date
1998-10-09
Last Version Date
2024-04-03
Entry Detail
PDB ID:
428D
Keywords:
Title:
STRUCTURE OF THE POTASSIUM FORM OF CGCGAATTCGCG: DNA DEFORMATION BY ELECTROSTATIC COLLAPSE AROUND SMALL CATIONS
Method Details:
Experimental Method:
Resolution:
1.75 Å
R-Value Free:
0.28
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21