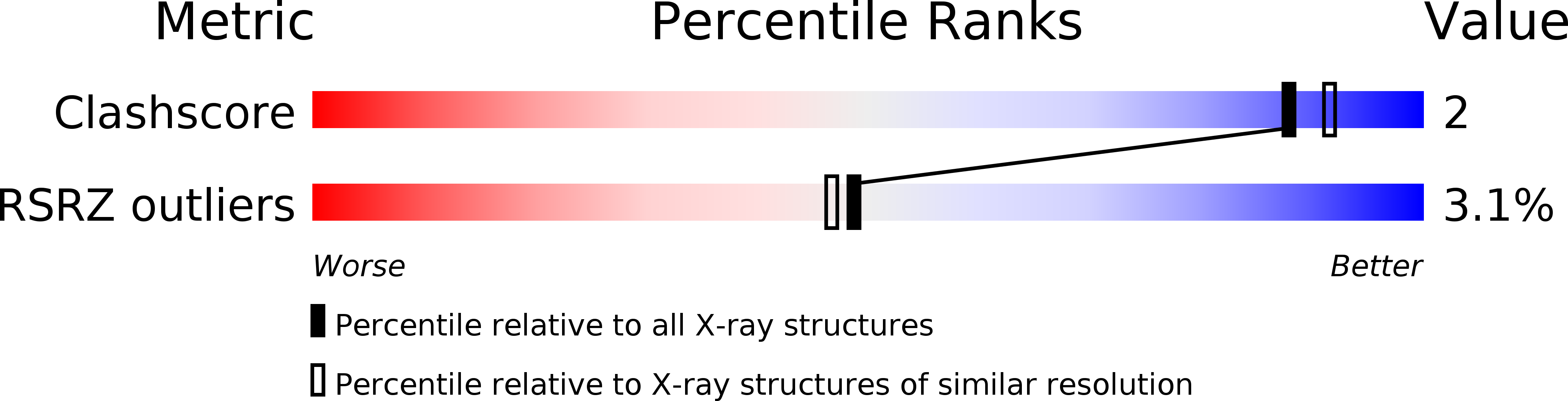Deposition Date
1998-08-12
Release Date
2000-05-29
Last Version Date
2024-04-03
Entry Detail
PDB ID:
419D
Keywords:
Title:
OCTAMER 5'-R(*GP*UP*AP*UP*AP*CP*A)-D(P*C)-3' WITH SIX WATSON-CRICK BASE-PAIRS AND TWO 3' OVERHANG RESIDUES
Method Details:
Experimental Method:
Resolution:
2.20 Å
R-Value Free:
0.27
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 21