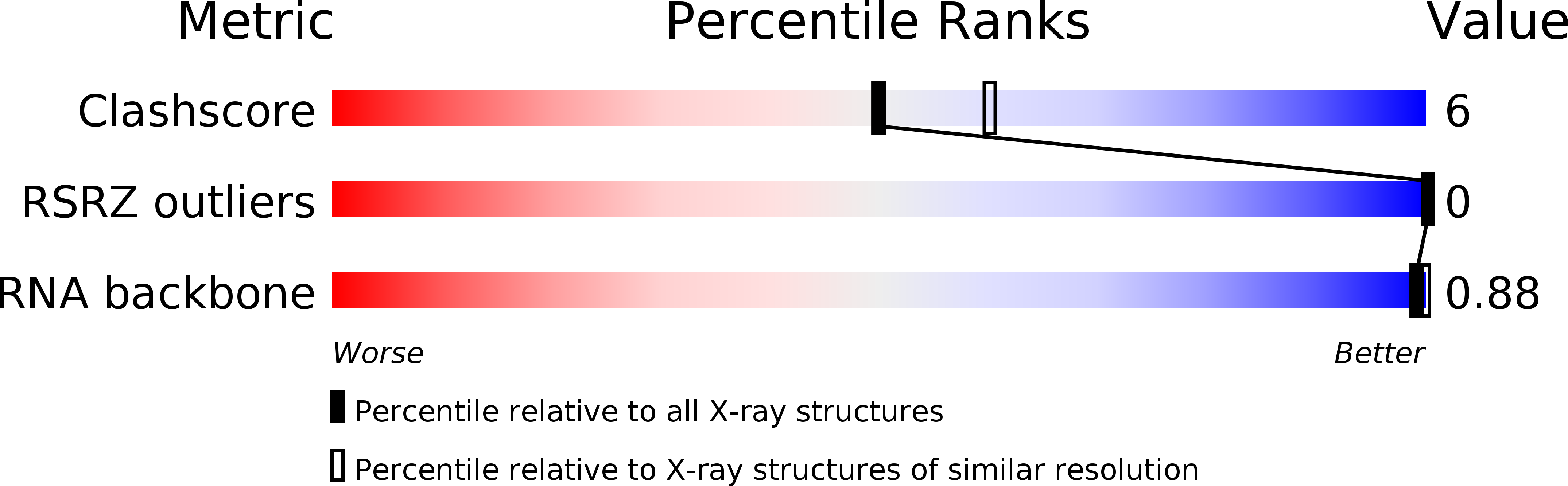
Deposition Date
1998-06-04
Release Date
1998-06-16
Last Version Date
2024-02-28
Entry Detail
Method Details:
Experimental Method:
Resolution:
2.30 Å
R-Value Free:
0.26
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
H 3 2