Deposition Date
2013-01-24
Release Date
2013-02-06
Last Version Date
2024-05-08
Entry Detail
PDB ID:
3ZKR
Keywords:
Title:
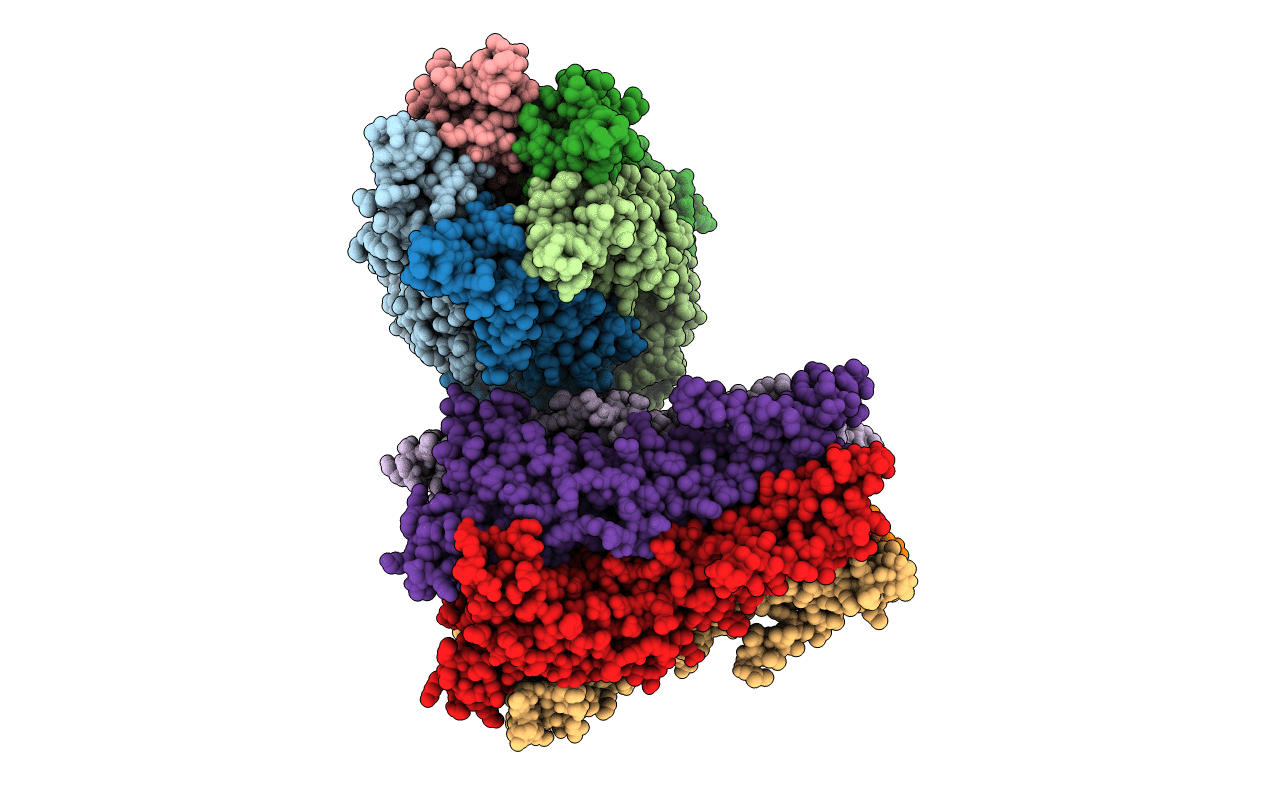
X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform
Biological Source:
Source Organism:
ERWINIA CHRYSANTHEMI (Taxon ID: 556)
Host Organism:
Method Details:
Experimental Method:
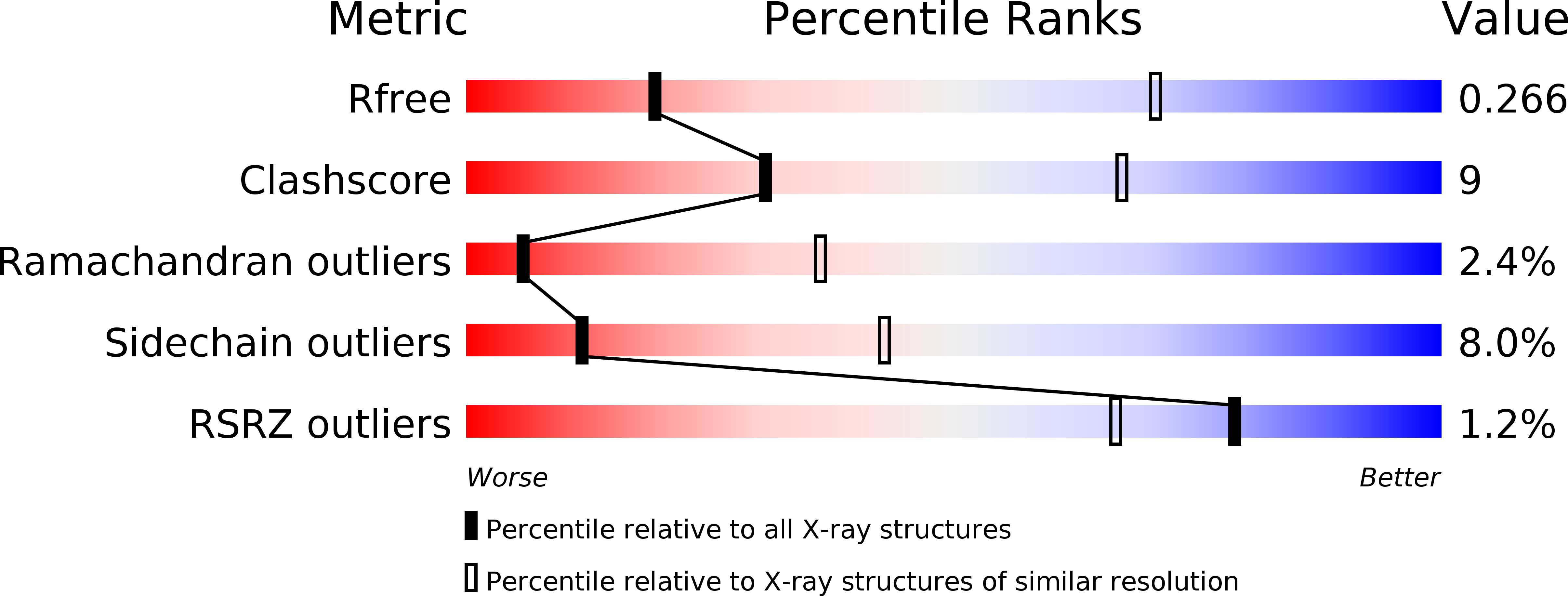
Resolution:
3.65 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
P 1 21 1