Deposition Date
2012-11-20
Release Date
2013-10-09
Last Version Date
2023-12-20
Entry Detail
PDB ID:
3ZCJ
Keywords:
Title:
Crystal structure of Helicobacter pylori T4SS protein CagL in a tetragonal crystal form with a helical RGD-motif (6 Mol per ASU)
Biological Source:
Source Organism(s):
HELICOBACTER PYLORI (Taxon ID: 85962)
Expression System(s):
Method Details:
Experimental Method:
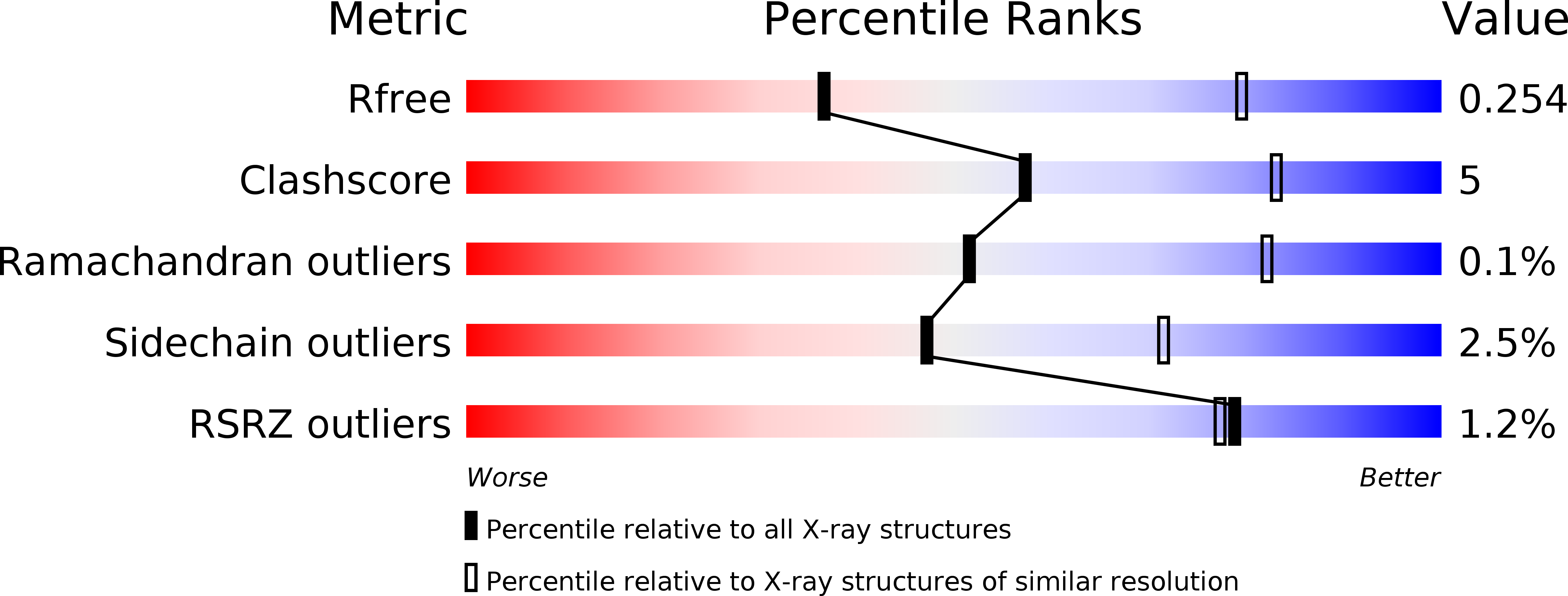
Resolution:
3.25 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 43 21 2