Deposition Date
2014-11-20
Release Date
2015-05-13
Last Version Date
2024-05-29
Entry Detail
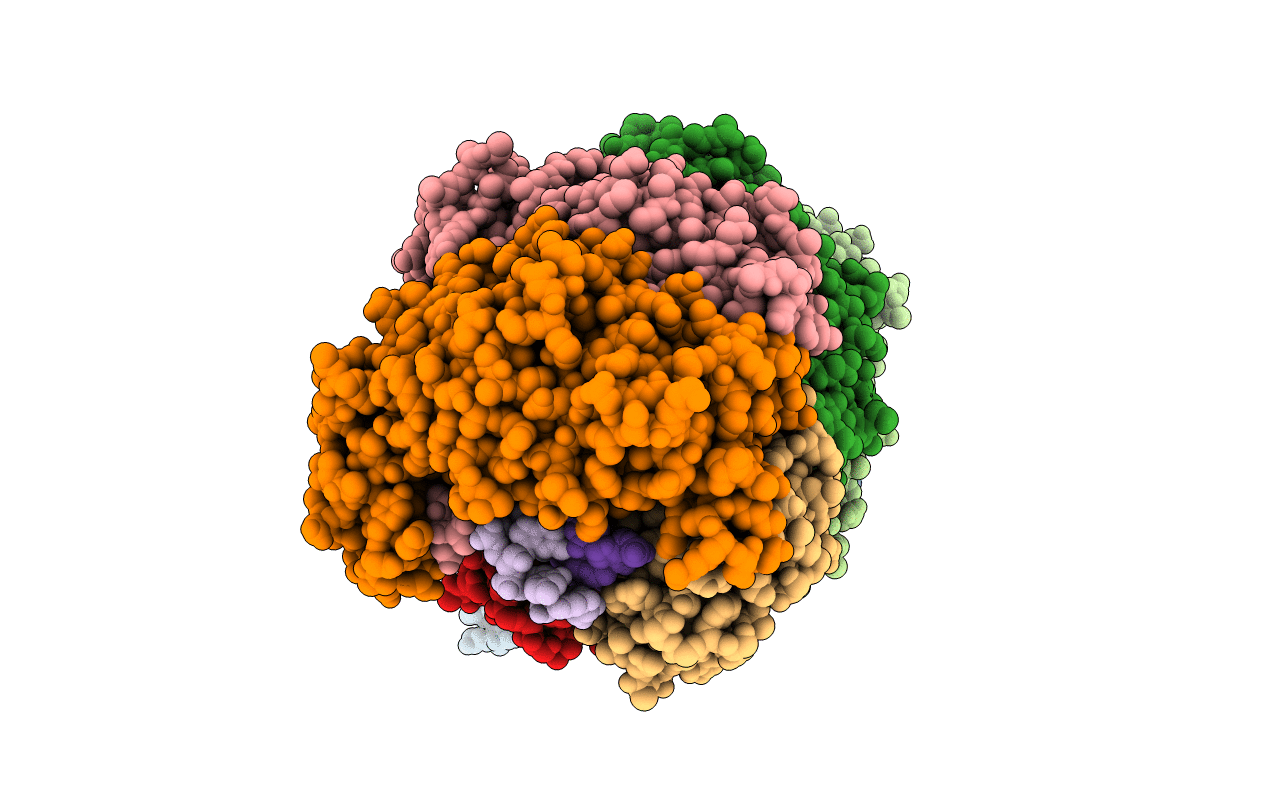
PDB ID:
3X1L
Keywords:
Title:
Crystal Structure of the CRISPR-Cas RNA Silencing Cmr Complex Bound to a Target Analog
Biological Source:
Source Organism(s):
Pyrococcus furiosus DSM 3638 (Taxon ID: 186497)
Archaeoglobus fulgidus DSM 4304 (Taxon ID: 224325)
Pyrococcus furiosus COM1 (Taxon ID: 1185654)
synthetic (Taxon ID: 32630)
Archaeoglobus fulgidus DSM 4304 (Taxon ID: 224325)
Pyrococcus furiosus COM1 (Taxon ID: 1185654)
synthetic (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.10 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.21
Space Group:
P 1


