Deposition Date
2011-12-21
Release Date
2012-06-06
Last Version Date
2024-10-30
Entry Detail
PDB ID:
3VN3
Keywords:
Title:
Fungal antifreeze protein exerts hyperactivity by constructing an inequable beta-helix
Biological Source:
Source Organism(s):
Typhula ishikariensis (Taxon ID: 69361)
Method Details:
Experimental Method:
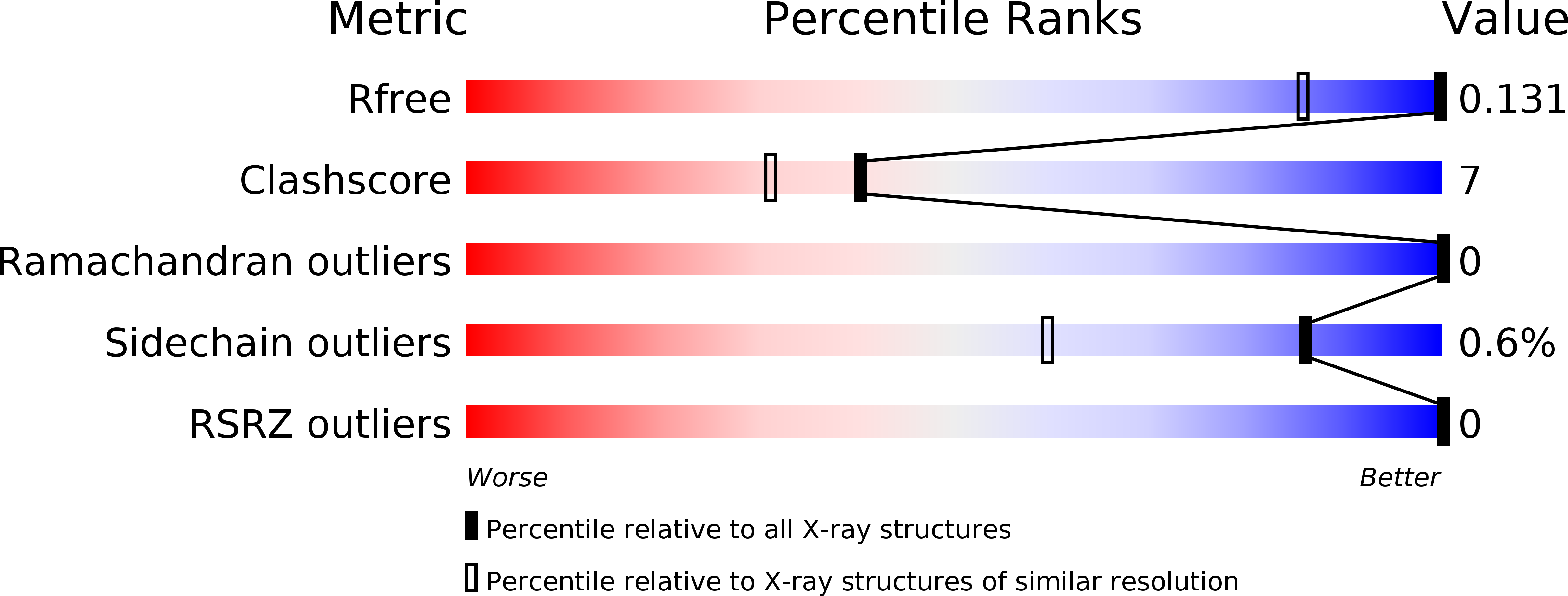
Resolution:
0.95 Å
R-Value Free:
0.12
R-Value Work:
0.11
R-Value Observed:
0.11
Space Group:
P 1