Deposition Date
1987-06-29
Release Date
1989-01-09
Last Version Date
2024-05-22
Entry Detail
PDB ID:
3TMN
Keywords:
Title:
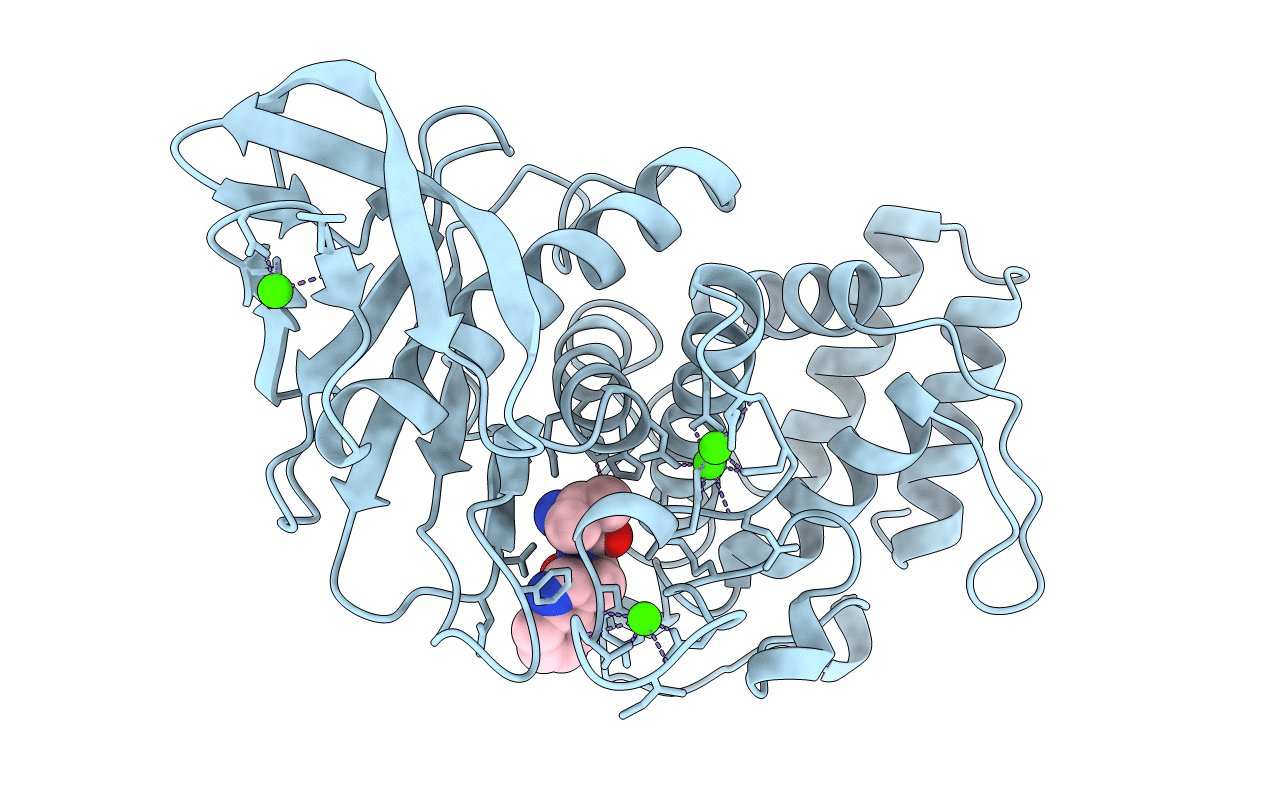
THE BINDING OF L-VALYL-L-TRYPTOPHAN TO CRYSTALLINE THERMOLYSIN ILLUSTRATES THE MODE OF INTERACTION OF A PRODUCT OF PEPTIDE HYDROLYSIS
Biological Source:
Source Organism(s):
Bacillus thermoproteolyticus (Taxon ID: 1427)
Method Details:
Experimental Method:
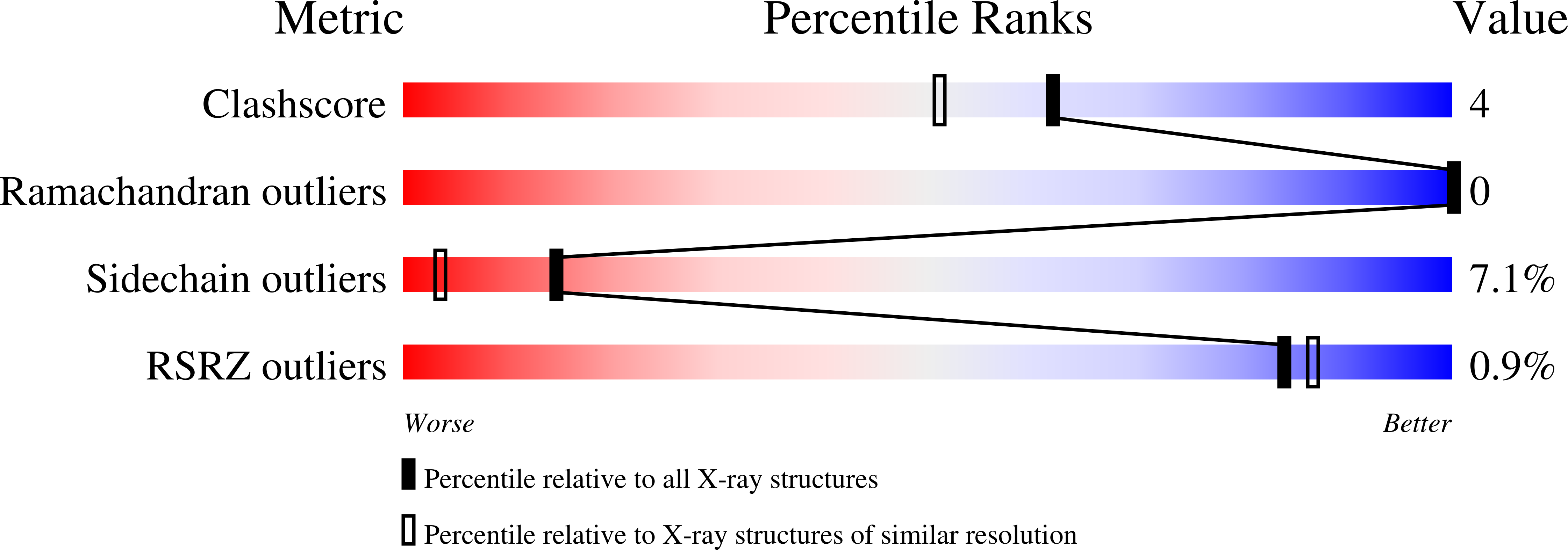
Resolution:
1.70 Å
R-Value Observed:
0.17
Space Group:
P 61 2 2