Deposition Date
2011-07-25
Release Date
2011-10-19
Last Version Date
2023-09-13
Entry Detail
PDB ID:
3T3N
Keywords:
Title:
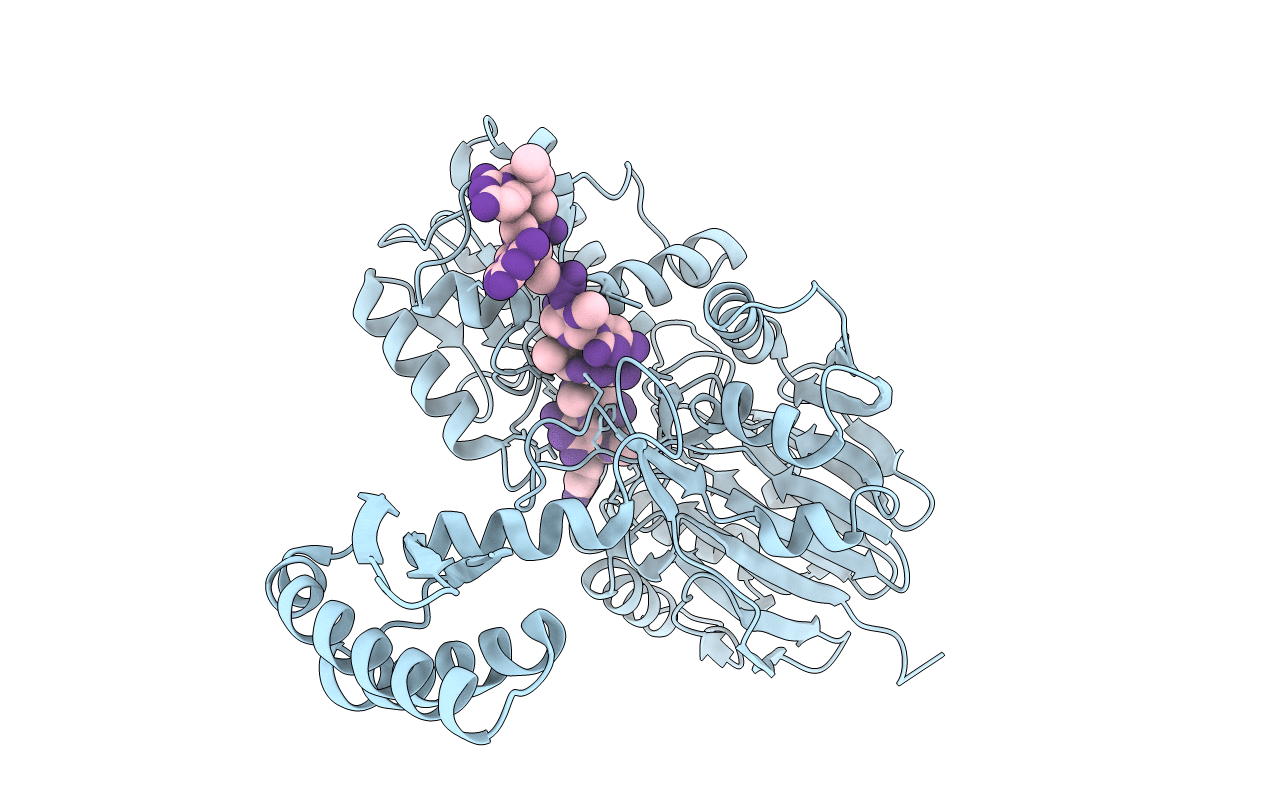
Molecular basis for the recognition and cleavage of RNA (UUCCGU) by the bifunctional 5'-3' exo/endoribonuclease RNase J
Biological Source:
Source Organism(s):
Thermus thermophilus HB27 (Taxon ID: 262724)
Expression System(s):
Method Details:
Experimental Method:
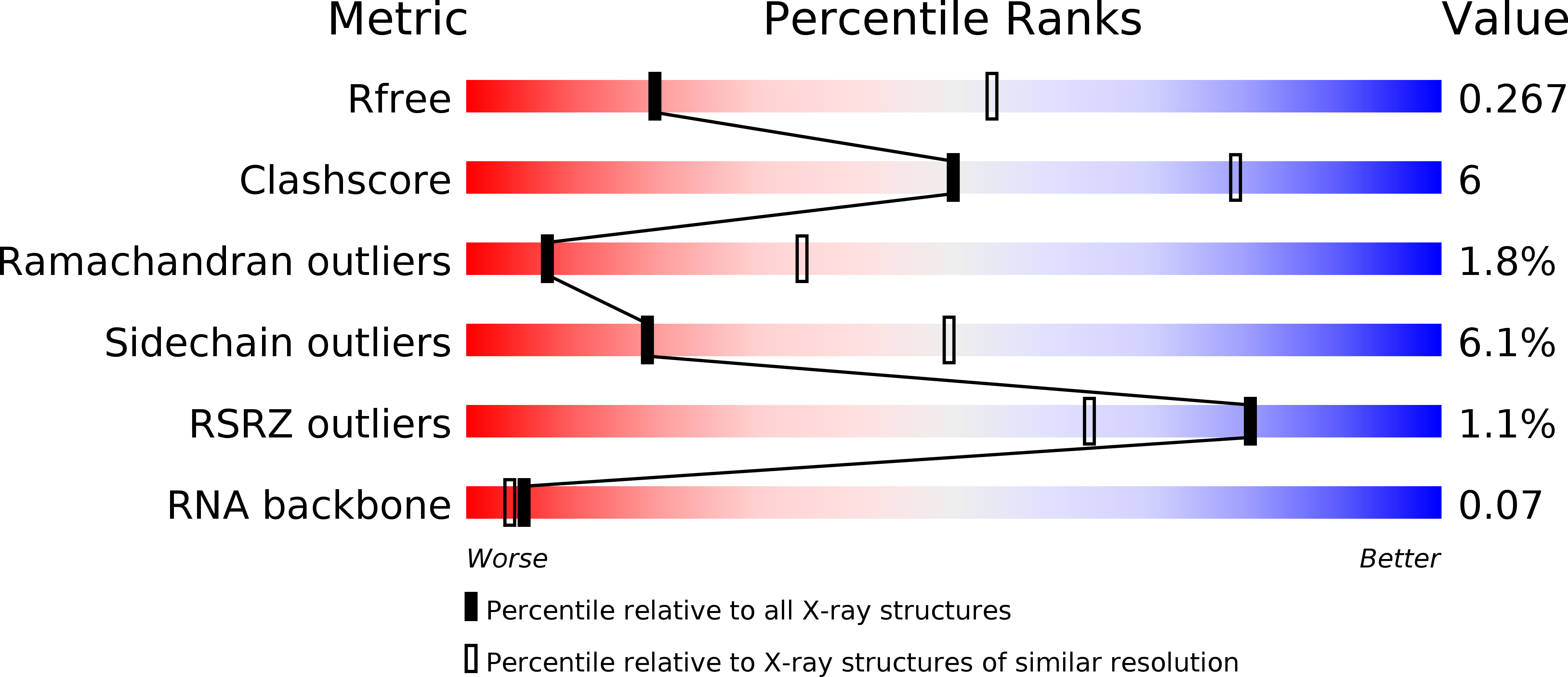
Resolution:
3.09 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
F 2 2 2