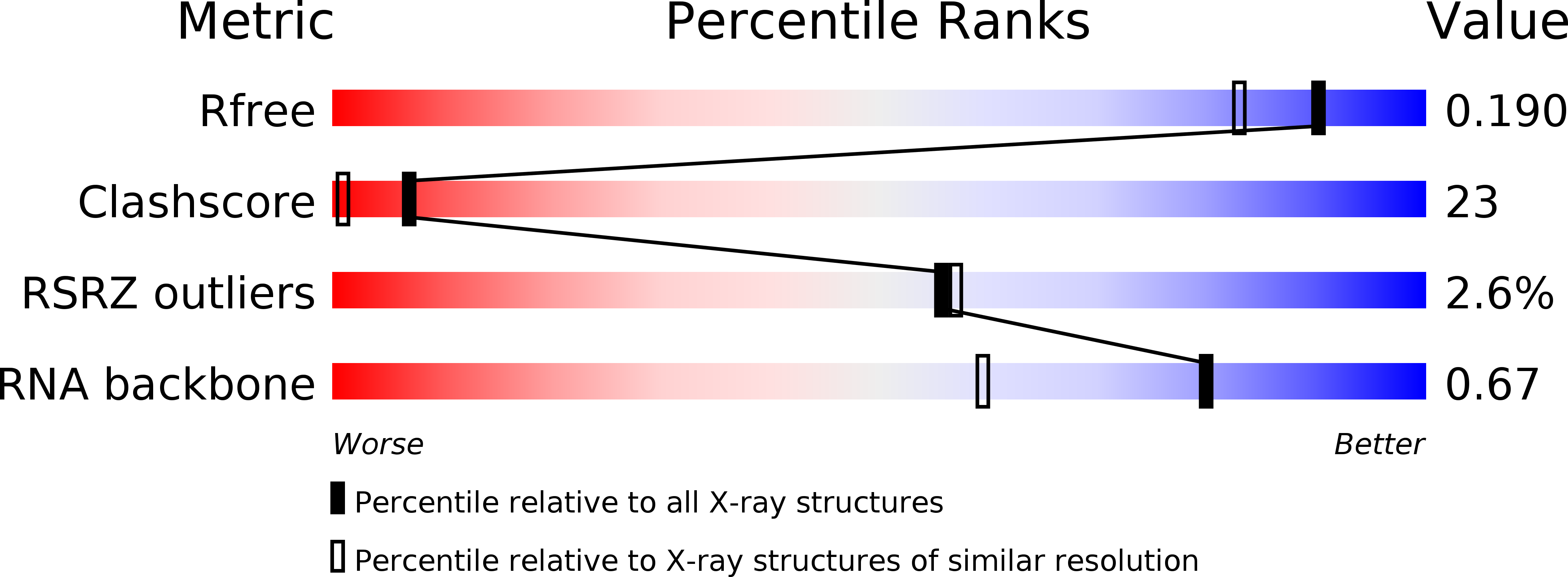Deposition Date
2011-07-18
Release Date
2012-04-25
Last Version Date
2024-03-13
Entry Detail
PDB ID:
3SYW
Keywords:
Title:
Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations
Method Details:
Experimental Method:
Resolution:
1.57 Å
R-Value Free:
0.18
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
H 3