Deposition Date
2011-07-14
Release Date
2012-05-16
Last Version Date
2024-10-09
Entry Detail
PDB ID:
3SX6
Keywords:
Title:
Crystal structure of sulfide:quinone oxidoreductase Cys356Ala variant from Acidithiobacillus ferrooxidans complexed with decylubiquinone
Biological Source:
Source Organism(s):
Acidithiobacillus ferrooxidans ATCC 23270 (Taxon ID: 243159)
Expression System(s):
Method Details:
Experimental Method:
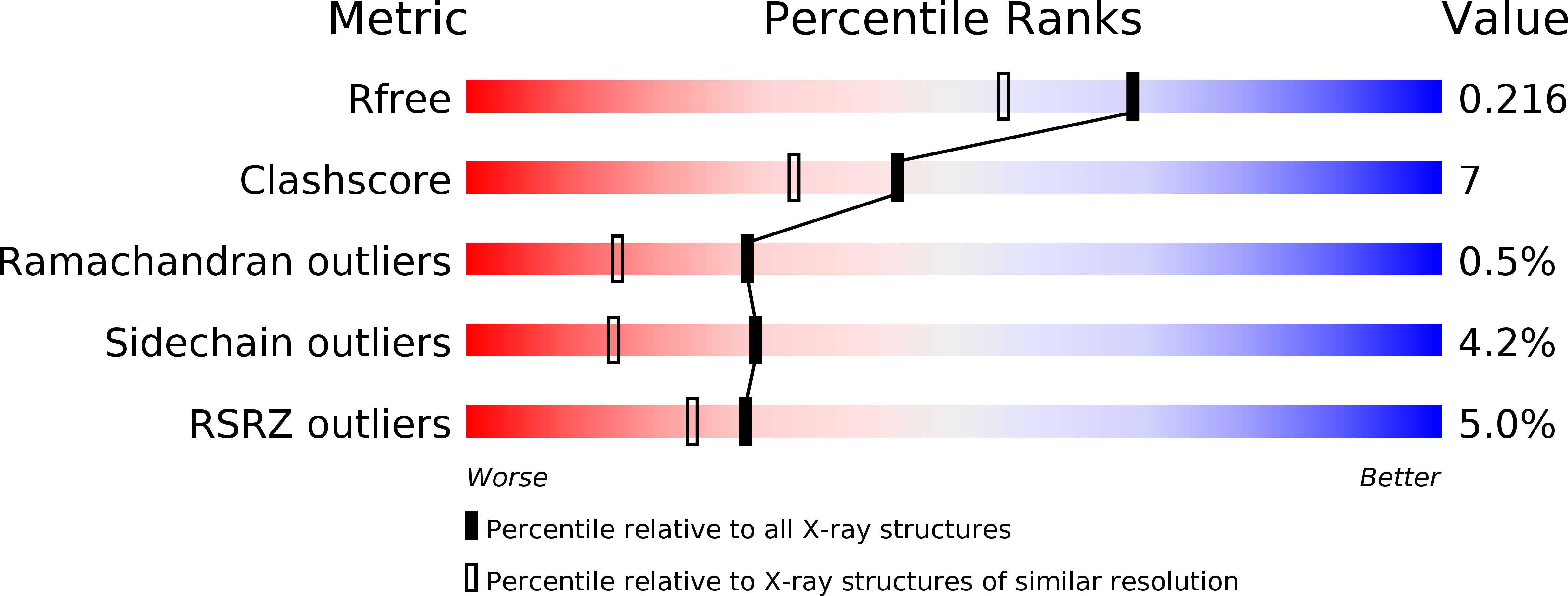
Resolution:
1.80 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.18
Space Group:
P 62 2 2