Deposition Date
2011-04-06
Release Date
2012-03-14
Last Version Date
2024-11-06
Entry Detail
PDB ID:
3RFJ
Keywords:
Title:
Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering
Biological Source:
Source Organism(s):
Listeria monocytogenes (Taxon ID: 637381)
Petromyzon marinus (Taxon ID: 7757)
synthetic (Taxon ID: 32630)
Petromyzon marinus (Taxon ID: 7757)
synthetic (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
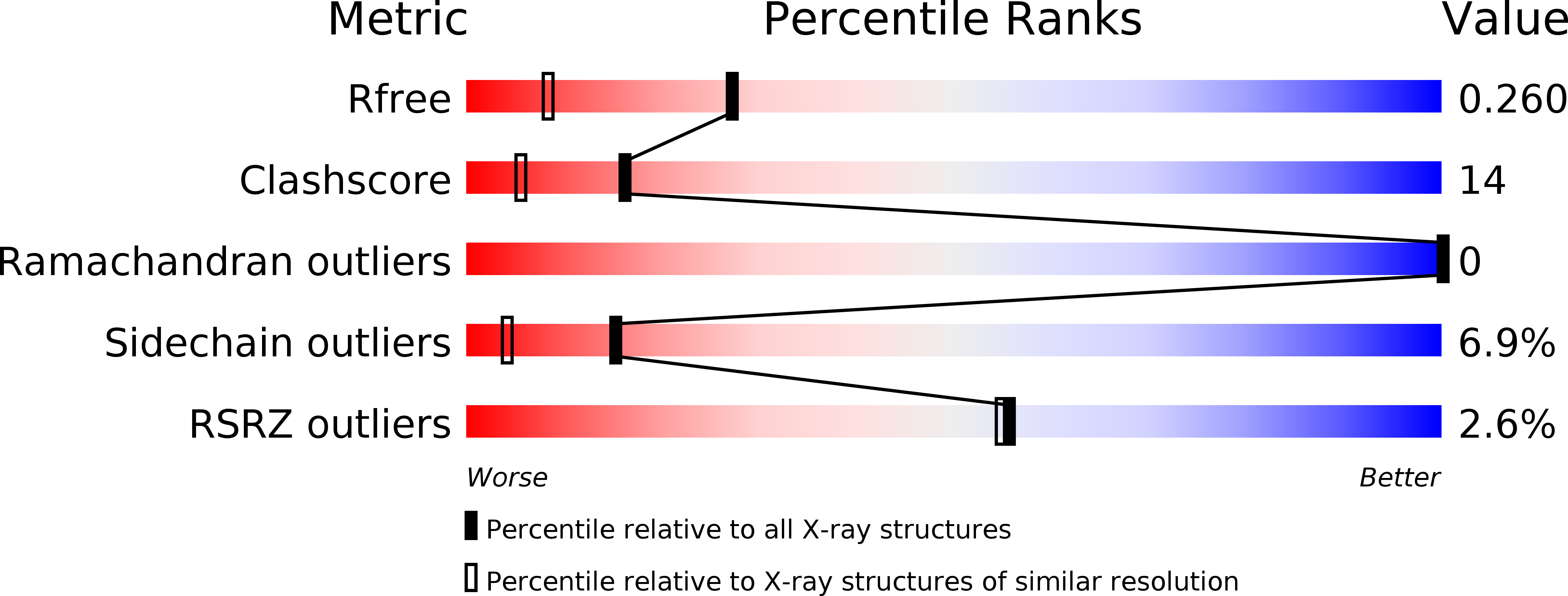
Resolution:
1.78 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 21 21 21