Deposition Date
2011-01-24
Release Date
2011-04-20
Last Version Date
2023-09-13
Entry Detail
PDB ID:
3QGH
Keywords:
Title:
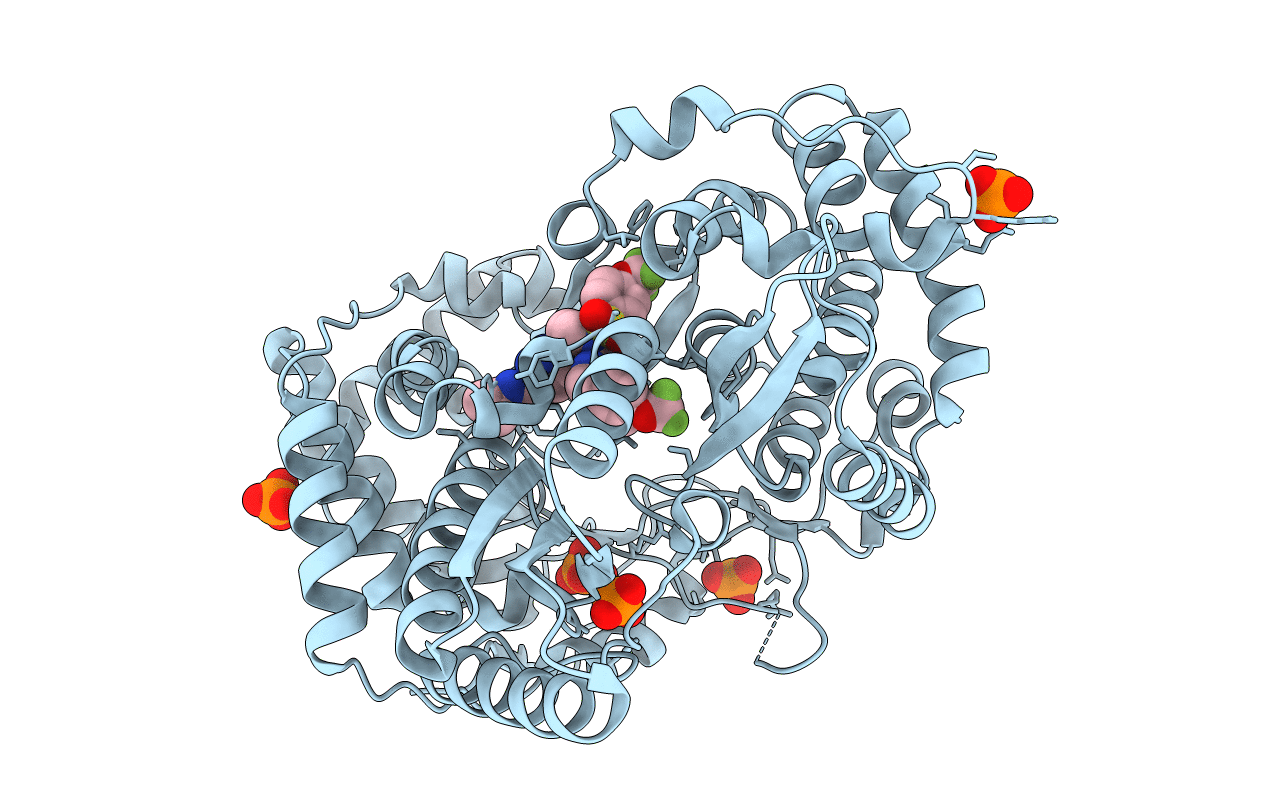
Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase genotype 1a complex with N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide
Biological Source:
Source Organism(s):
Hepatitis C virus subtype 1a (Taxon ID: 31646)
Expression System(s):
Method Details:
Experimental Method:
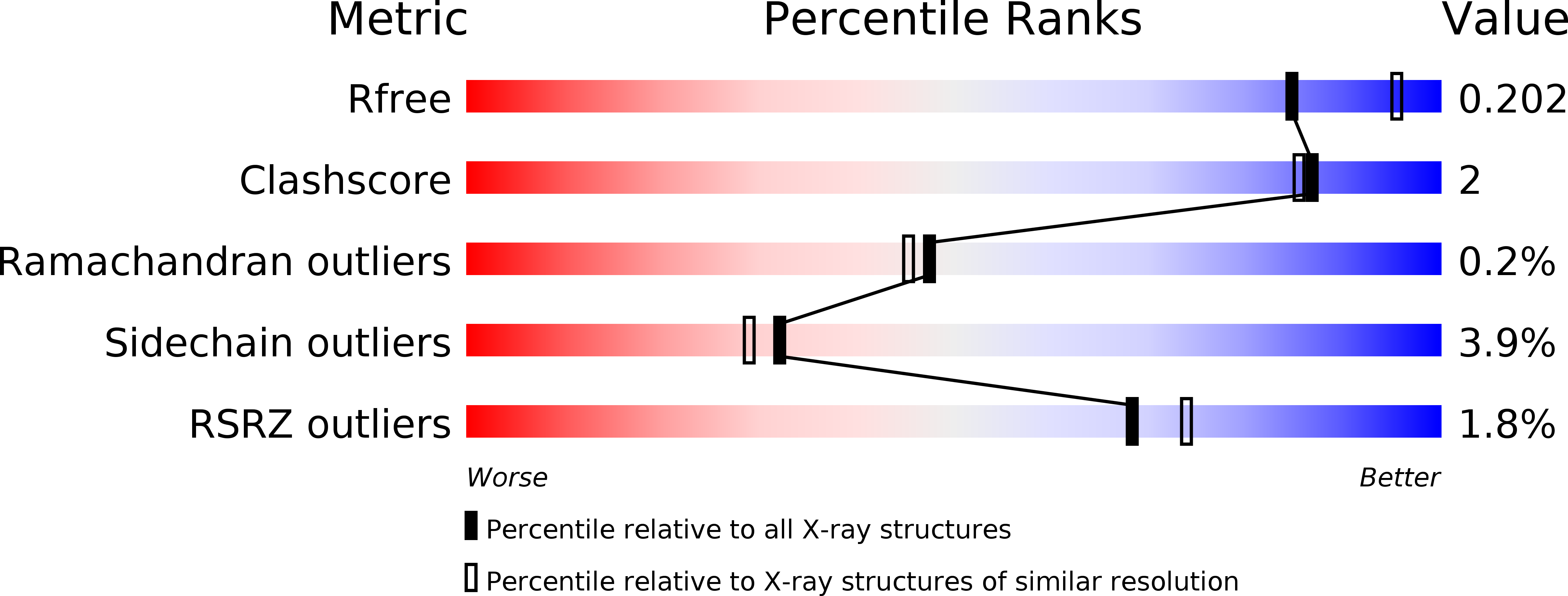
Resolution:
2.14 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21