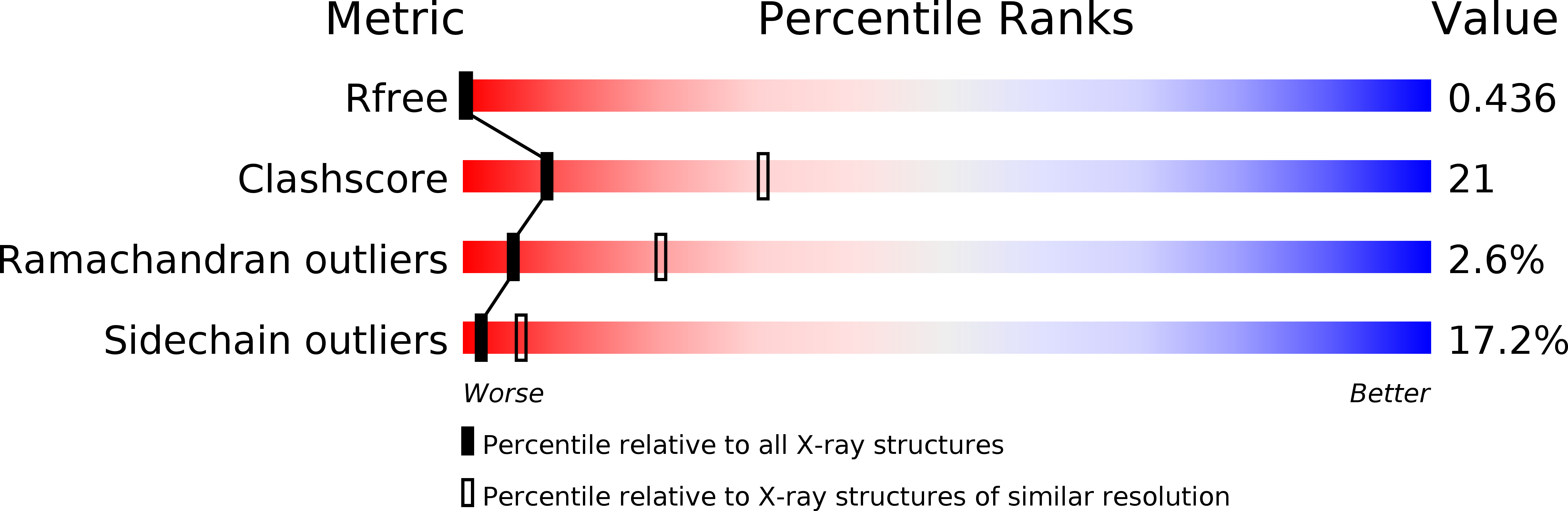
Deposition Date
2011-01-15
Release Date
2011-05-25
Last Version Date
2024-03-20
Entry Detail
PDB ID:
3QCA
Keywords:
Title:
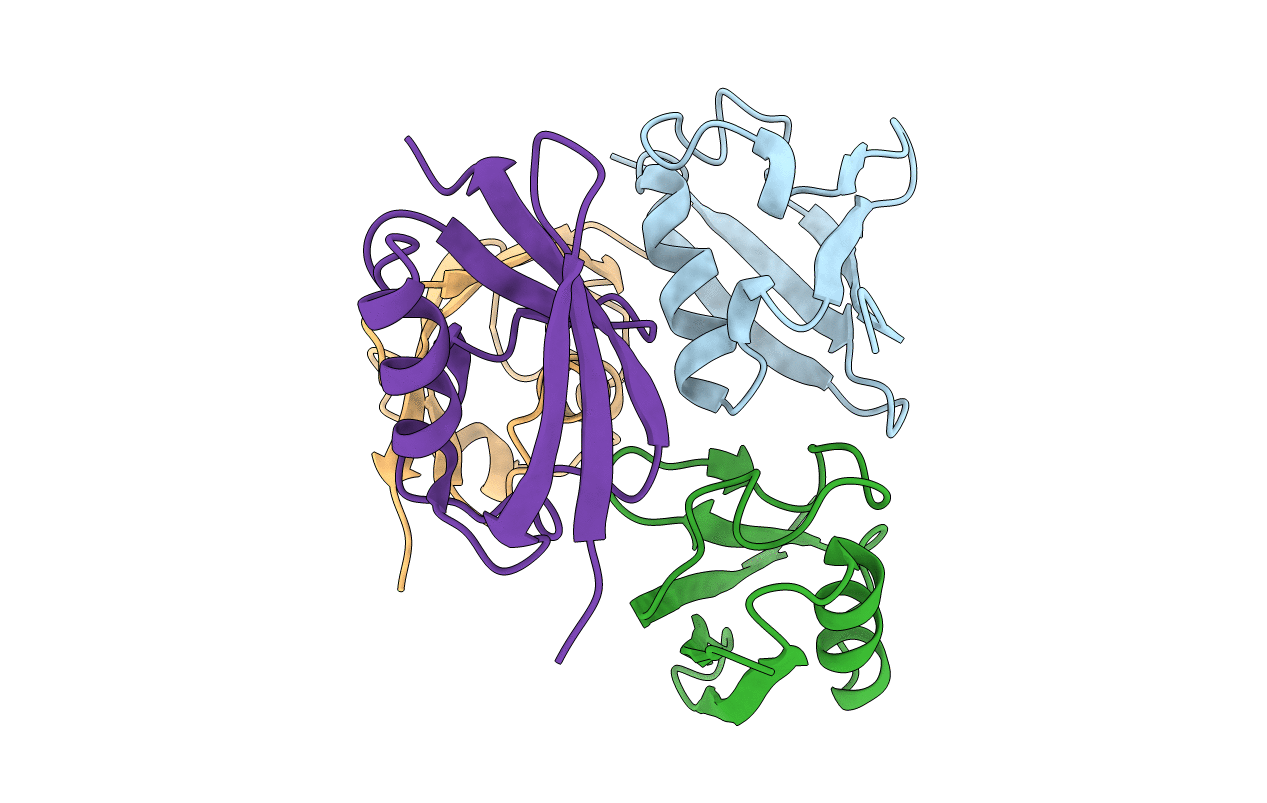
Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.90 Å
R-Value Free:
0.29
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
F 2 3