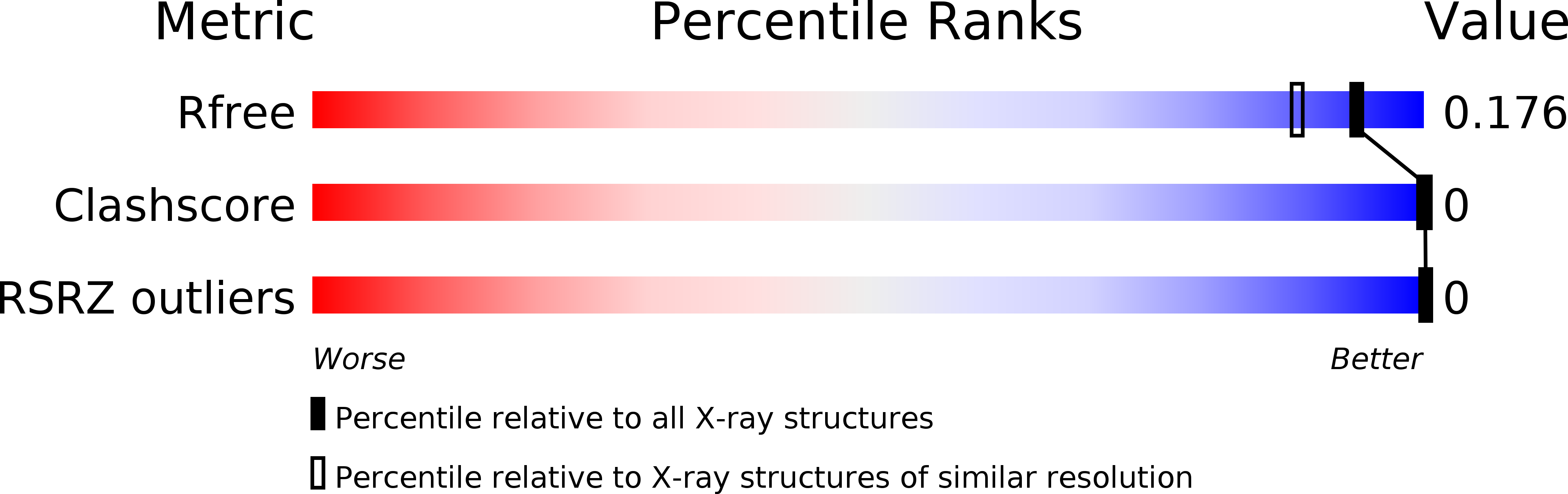Deposition Date
2011-01-12
Release Date
2011-03-02
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3QBA
Keywords:
Title:
Reintroducing Electrostatics into Macromolecular Crystallographic Refinement: Z-DNA (X-ray)
Method Details:
Experimental Method:
R-Value Free:
['0.23
R-Value Work:
['0.19
R-Value Observed:
['?', '?'].00
Space Group:
P 21 21 21