Deposition Date
2010-12-16
Release Date
2011-04-27
Last Version Date
2023-09-13
Entry Detail
PDB ID:
3Q0Z
Keywords:
Title:
Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid
Biological Source:
Source Organism(s):
Hepatitis C virus subtype 1b (Taxon ID: 31647)
Expression System(s):
Method Details:
Experimental Method:
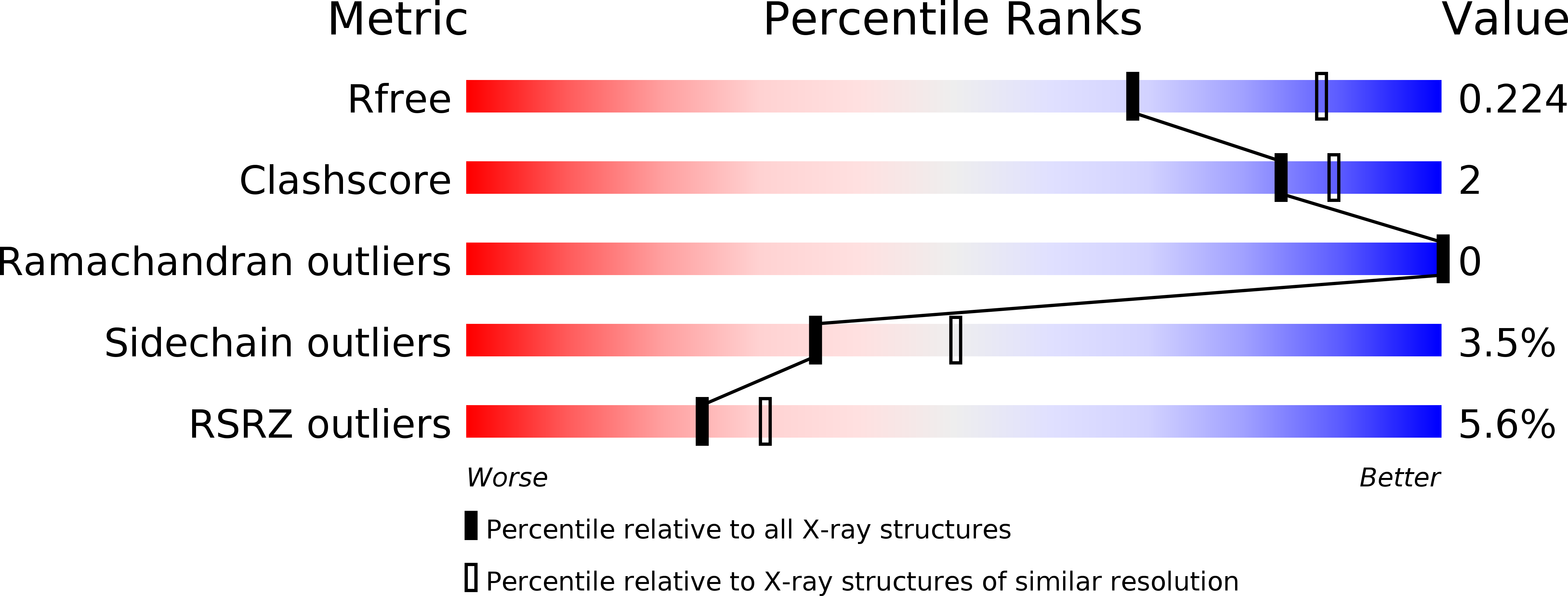
Resolution:
2.29 Å
R-Value Free:
0.22
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21