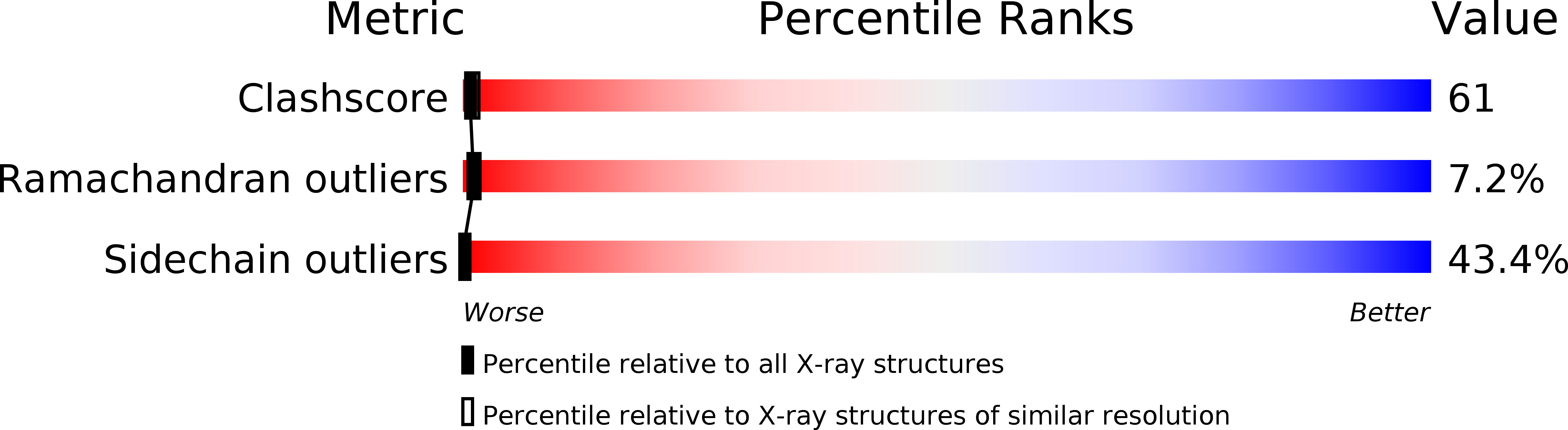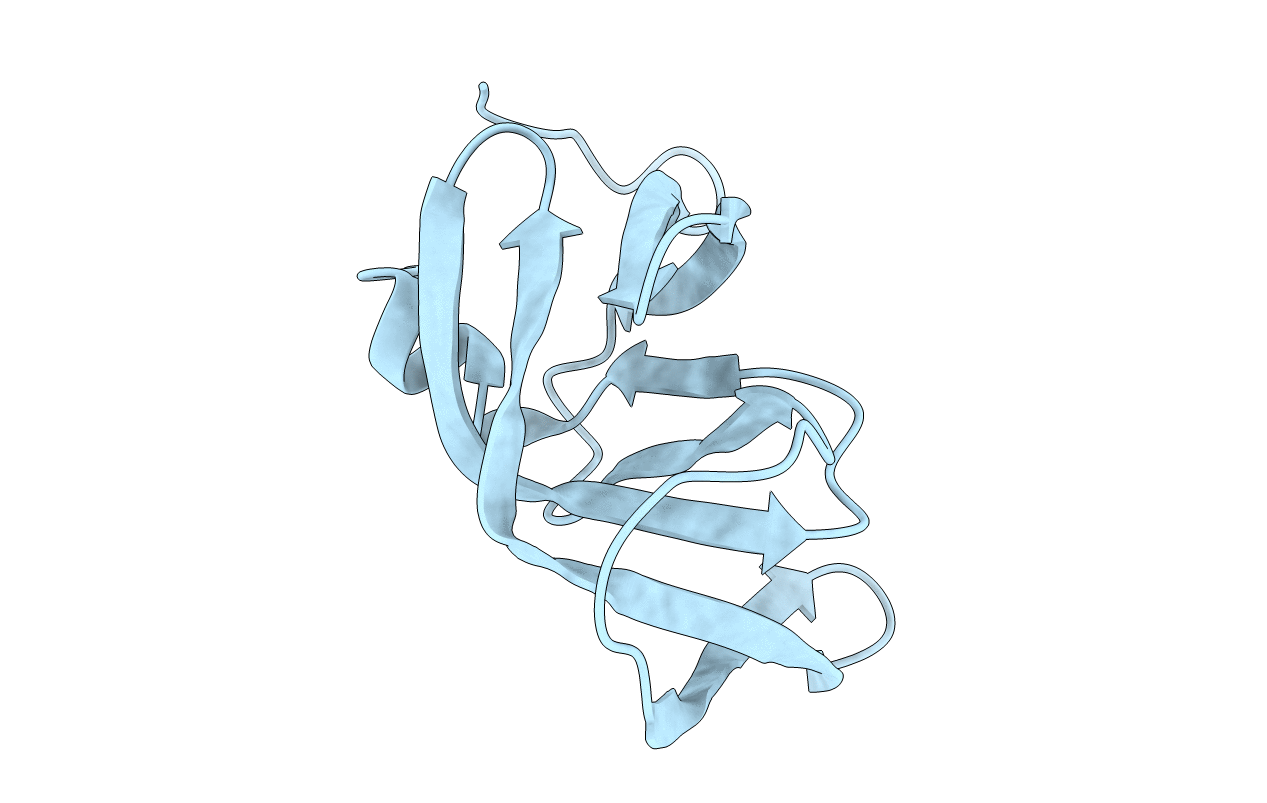
Deposition Date
1991-11-04
Release Date
1992-01-15
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3PHV
Keywords:
Title:
X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES
Biological Source:
Source Organism(s):
HIV-1 M:B_HXB2R (Taxon ID: 11706)
Expression System(s):
Method Details: