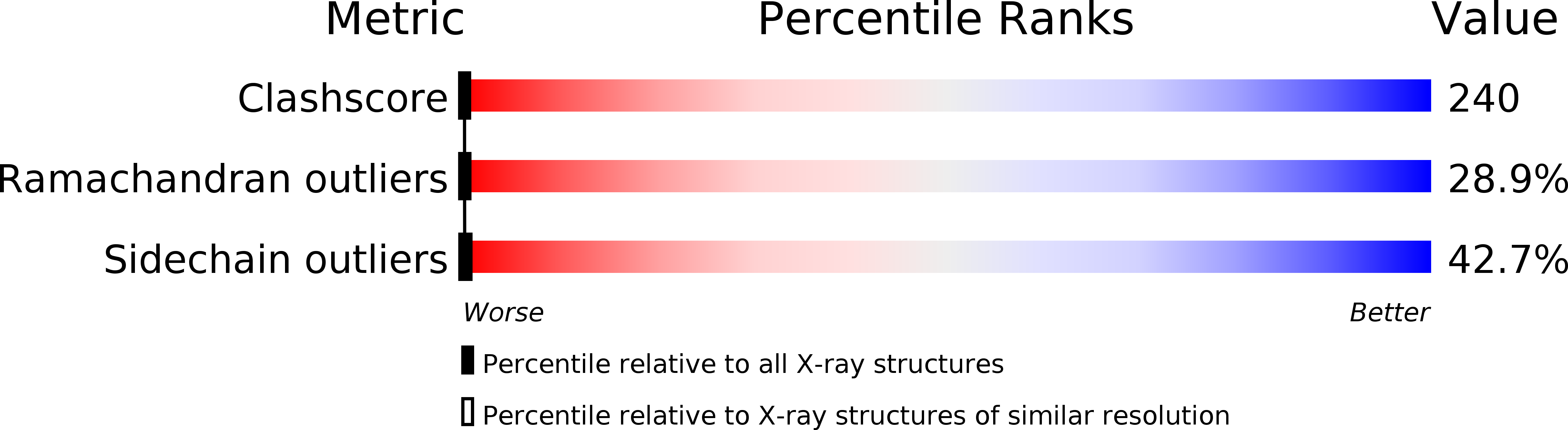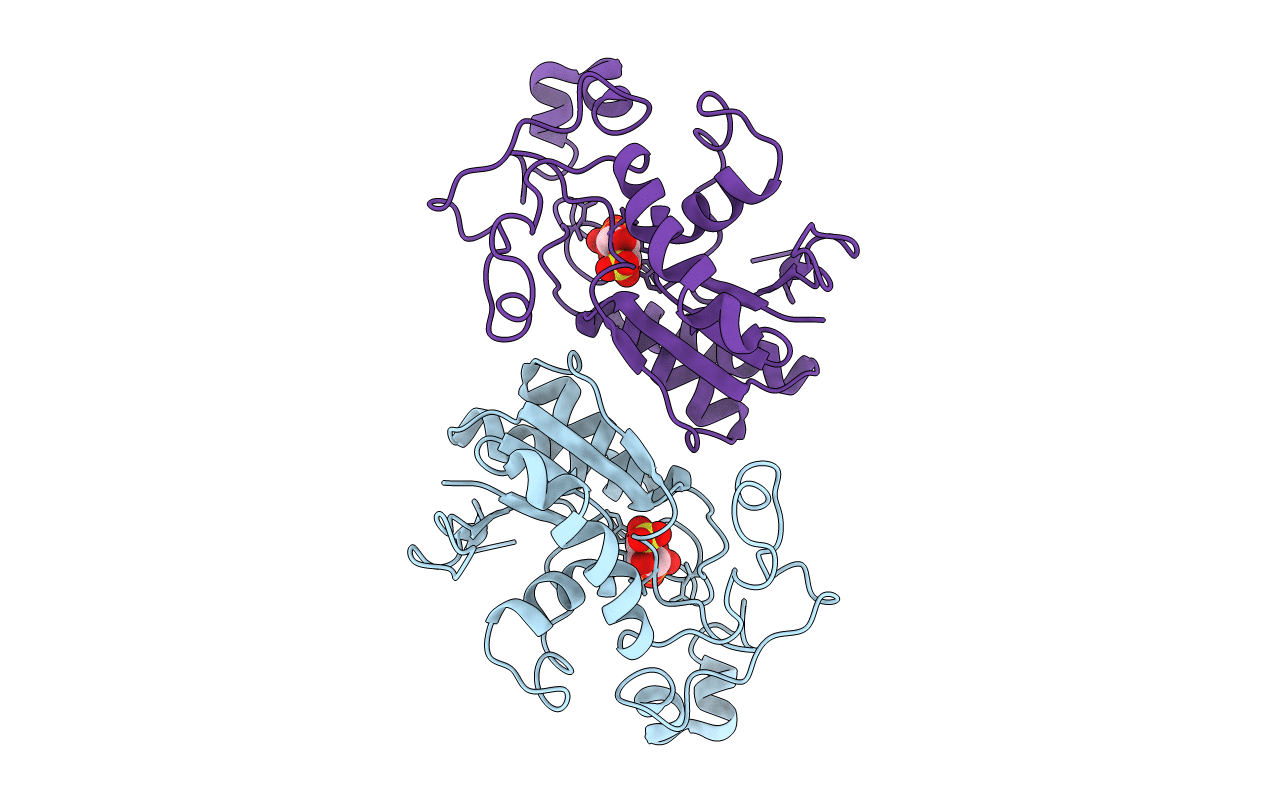
Deposition Date
1982-04-06
Release Date
1982-05-26
Last Version Date
2024-05-22
Entry Detail
PDB ID:
3PGM
Keywords:
Title:
THE STRUCTURE OF YEAST PHOSPHOGLYCERATE MUTASE AT 0.28 NM RESOLUTION
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Method Details: