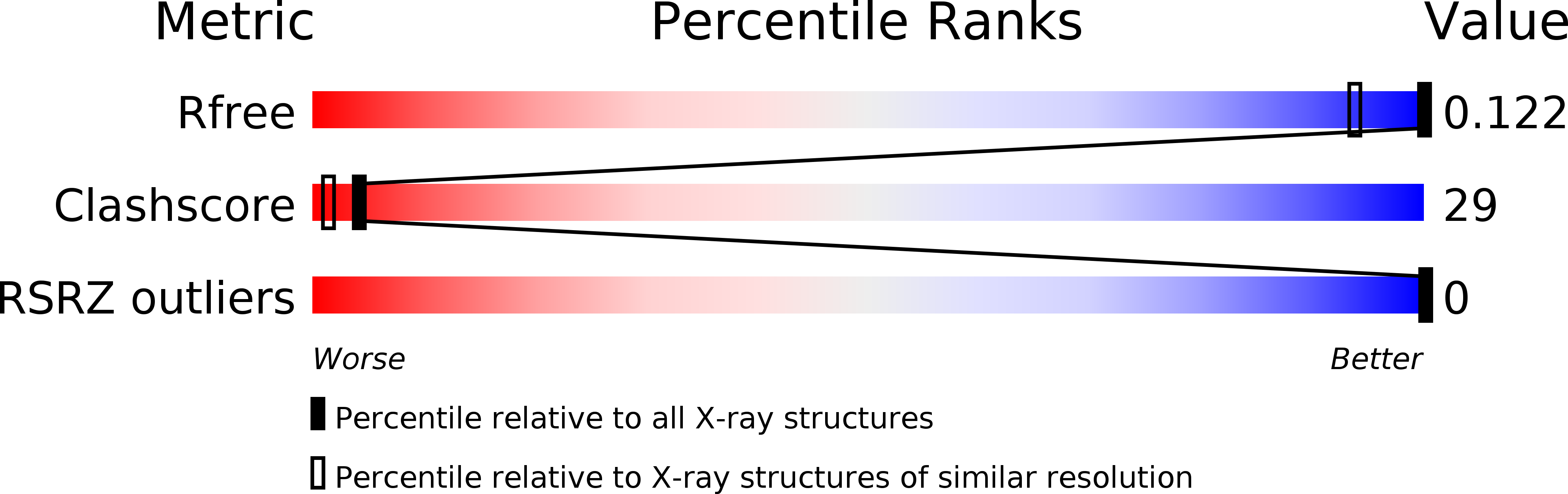
Deposition Date
2010-08-27
Release Date
2010-09-08
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3OMJ
Keywords:
Title:
Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding
Method Details:
Experimental Method:
Resolution:
0.95 Å
R-Value Free:
0.12
R-Value Work:
0.11
R-Value Observed:
0.11
Space Group:
P 41 21 2