Deposition Date
2010-07-22
Release Date
2010-08-04
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3O1X
Keywords:
Title:
High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C84A mutant from rabbit complexed with adenosine
Biological Source:
Source Organism:
Oryctolagus cuniculus (Taxon ID: 9986)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.08 Å
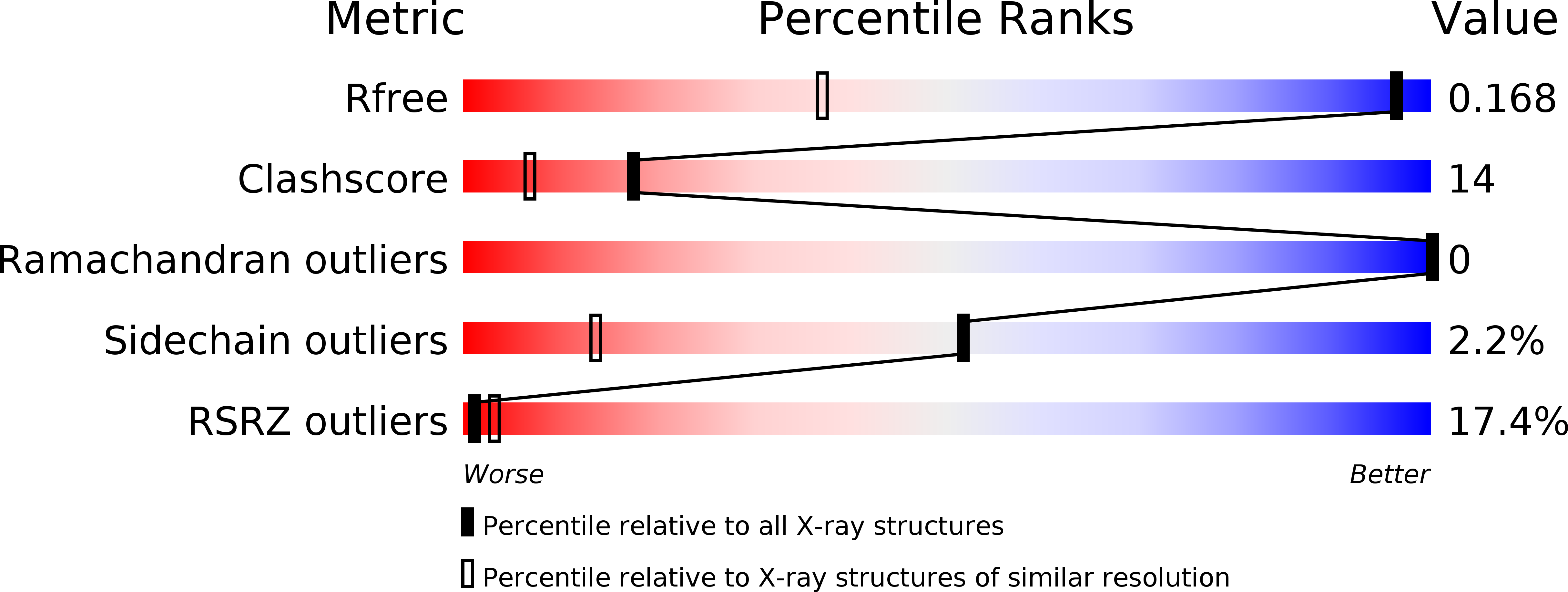
R-Value Free:
0.17
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 43 21 2