Deposition Date
2010-07-13
Release Date
2011-11-02
Last Version Date
2025-03-26
Entry Detail
PDB ID:
3NXE
Keywords:
Title:
X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with MVT-101 inhibitor
Method Details:
Experimental Method:
Resolution:
1.61 Å
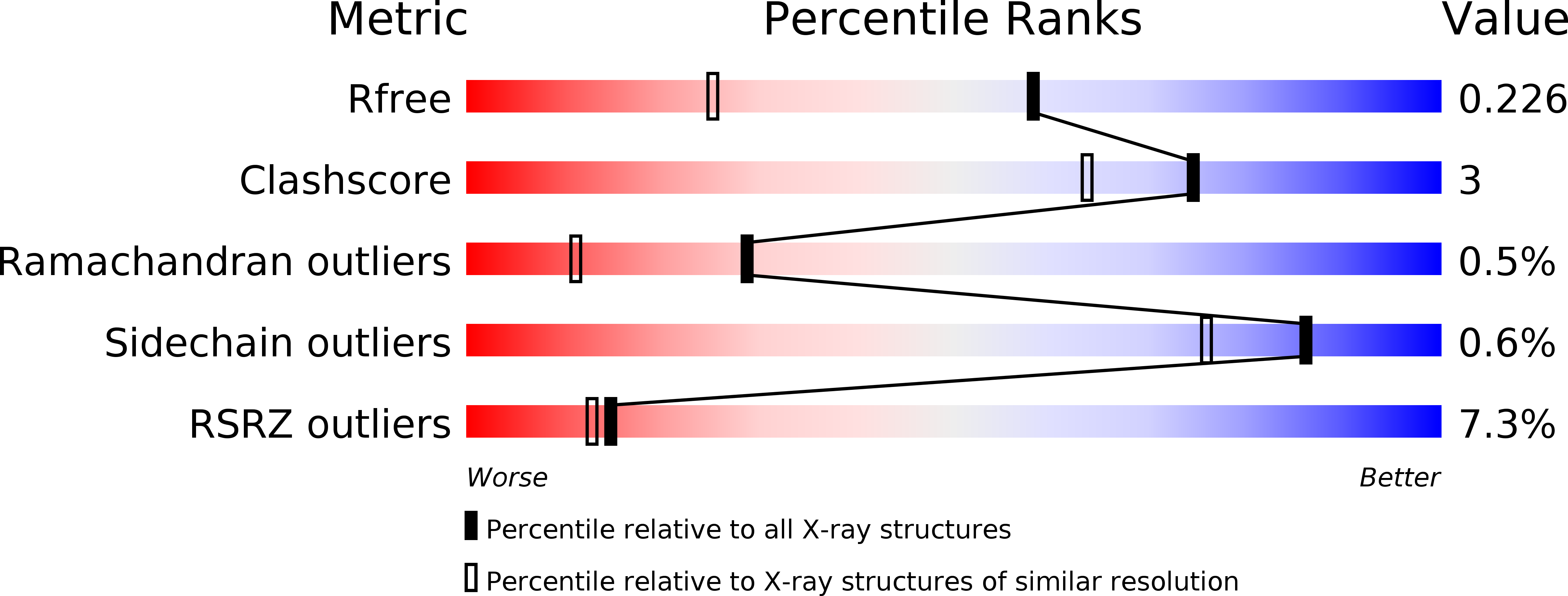
R-Value Free:
0.21
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 21