Deposition Date
2010-06-07
Release Date
2011-02-23
Last Version Date
2024-03-13
Entry Detail
PDB ID:
3NDB
Keywords:
Title:
Crystal structure of a signal sequence bound to the signal recognition particle
Biological Source:
Source Organism(s):
Methanocaldococcus jannaschii (Taxon ID: 2190)
Methanocaldococcus jannaschii DSM 2661 (Taxon ID: 243232)
Methanocaldococcus jannaschii DSM 2661 (Taxon ID: 243232)
Expression System(s):
Method Details:
Experimental Method:
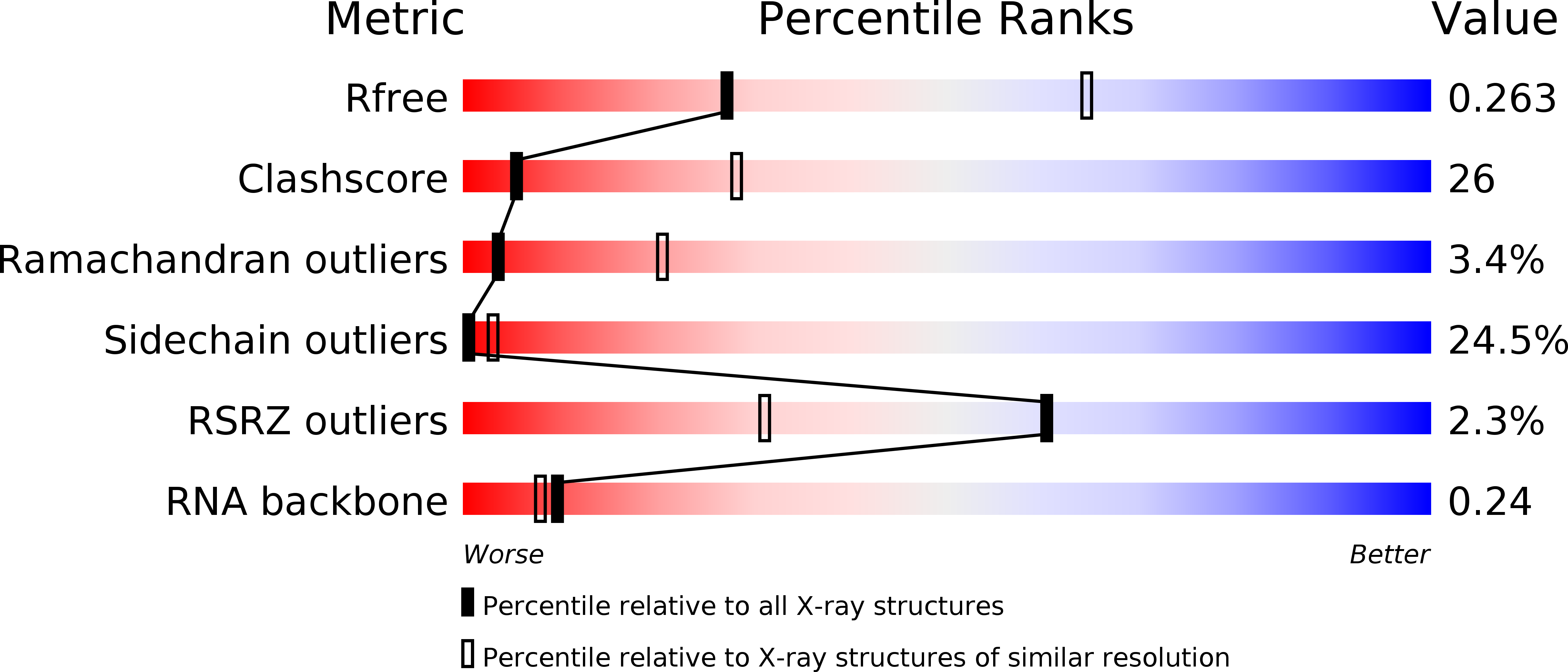
Resolution:
3.00 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
I 2 2 2