Deposition Date
2010-05-28
Release Date
2011-06-08
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3N97
Keywords:
Title:
RNA polymerase alpha C-terminal domain (E. coli) and sigma region 4 (T. aq. mutant) bound to (UP,-35 element) DNA
Biological Source:
Source Organism(s):
Thermus aquaticus (Taxon ID: 271)
Escherichia coli (Taxon ID: 83333)
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
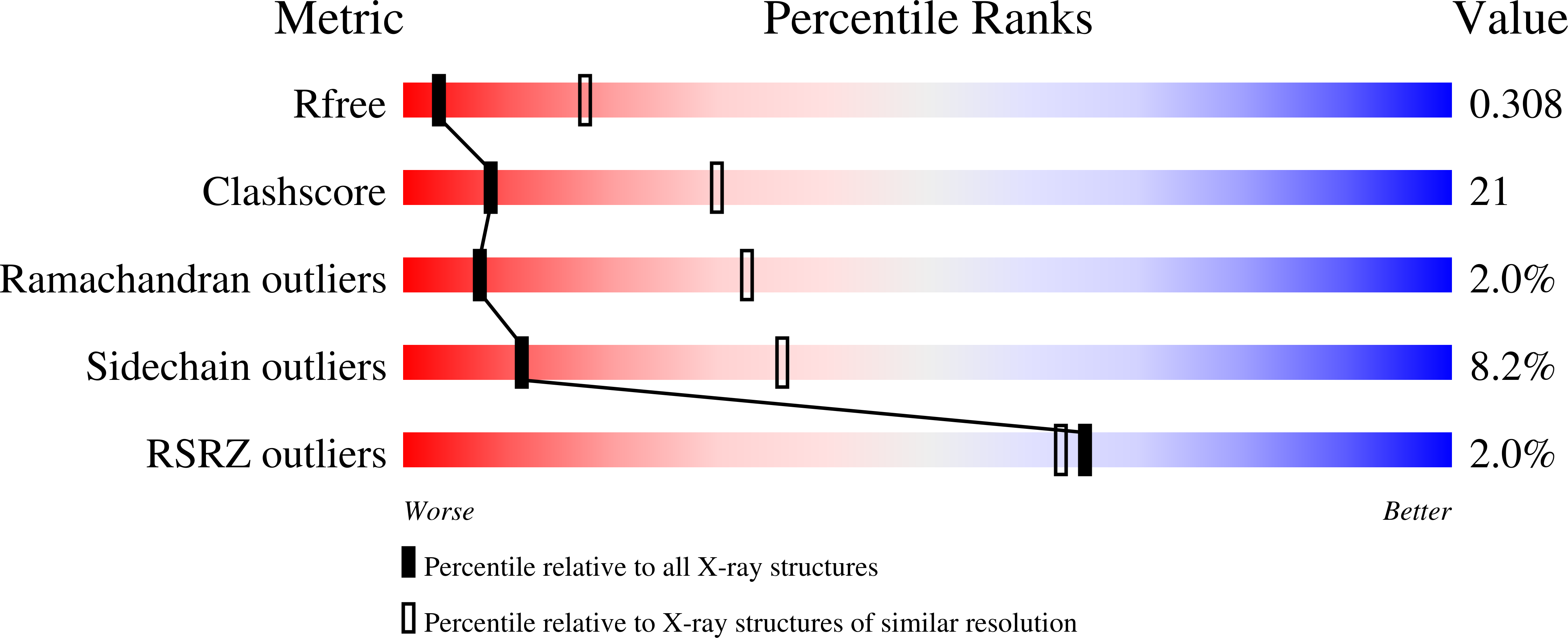
Resolution:
3.25 Å
R-Value Free:
0.29
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
C 2 2 21