Deposition Date
2010-04-30
Release Date
2011-01-05
Last Version Date
2023-11-22
Entry Detail
PDB ID:
3MT7
Keywords:
Title:
Glycogen phosphorylase complexed with 4-bromobenzaldehyde-4-(beta-D-glucopyranosyl)-thiosemicarbazone
Biological Source:
Source Organism(s):
Oryctolagus cuniculus (Taxon ID: 9986)
Method Details:
Experimental Method:
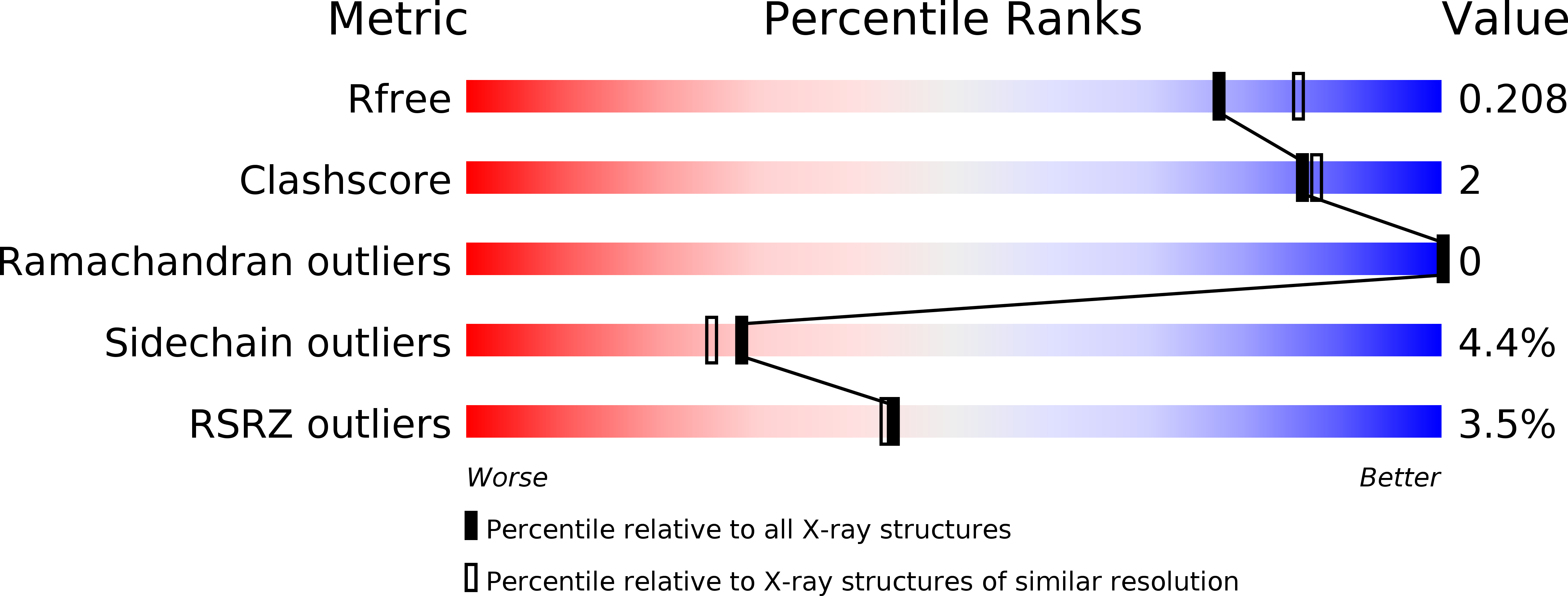
Resolution:
2.00 Å
R-Value Free:
0.20
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 43 21 2