Deposition Date
2010-04-12
Release Date
2010-06-30
Last Version Date
2024-11-20
Entry Detail
PDB ID:
3MJ5
Keywords:
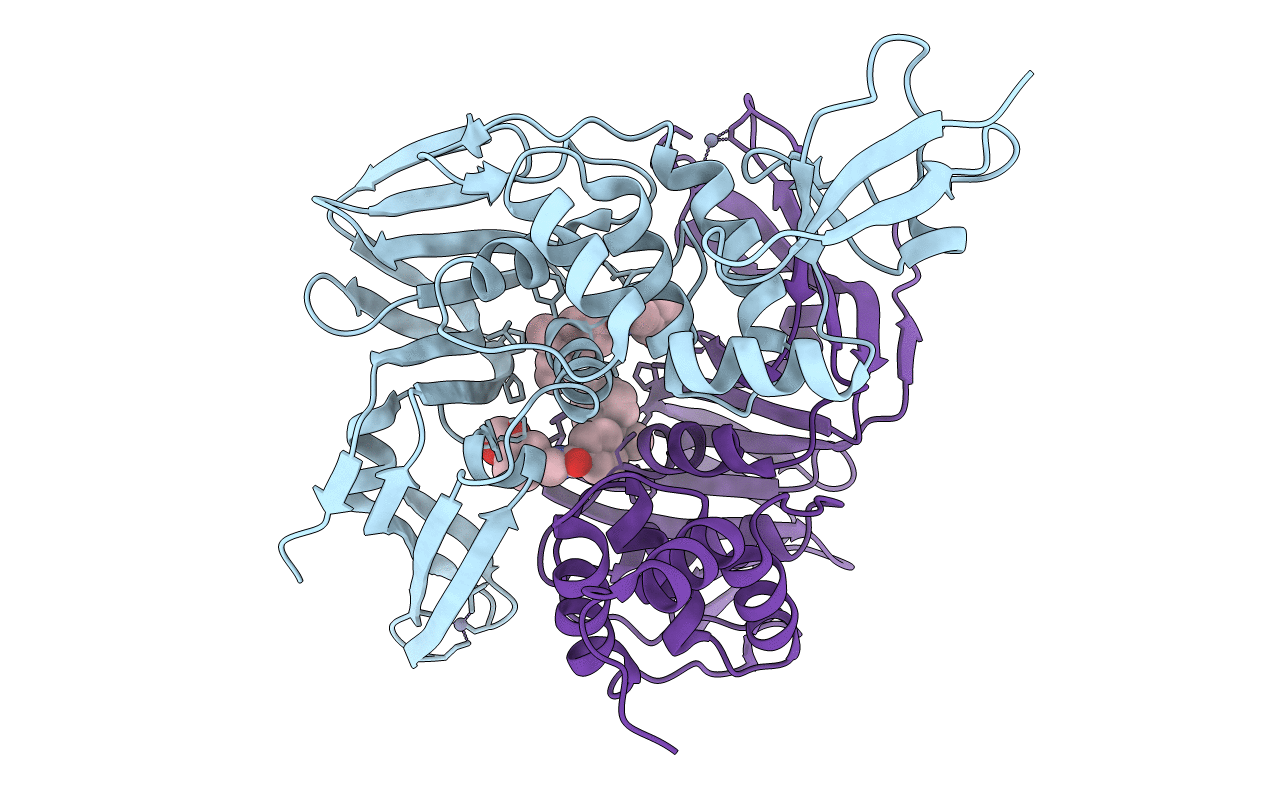
Title:
Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation
Biological Source:
Source Organism(s):
SARS coronavirus (Taxon ID: 227859)
Expression System(s):
Method Details:
Experimental Method:
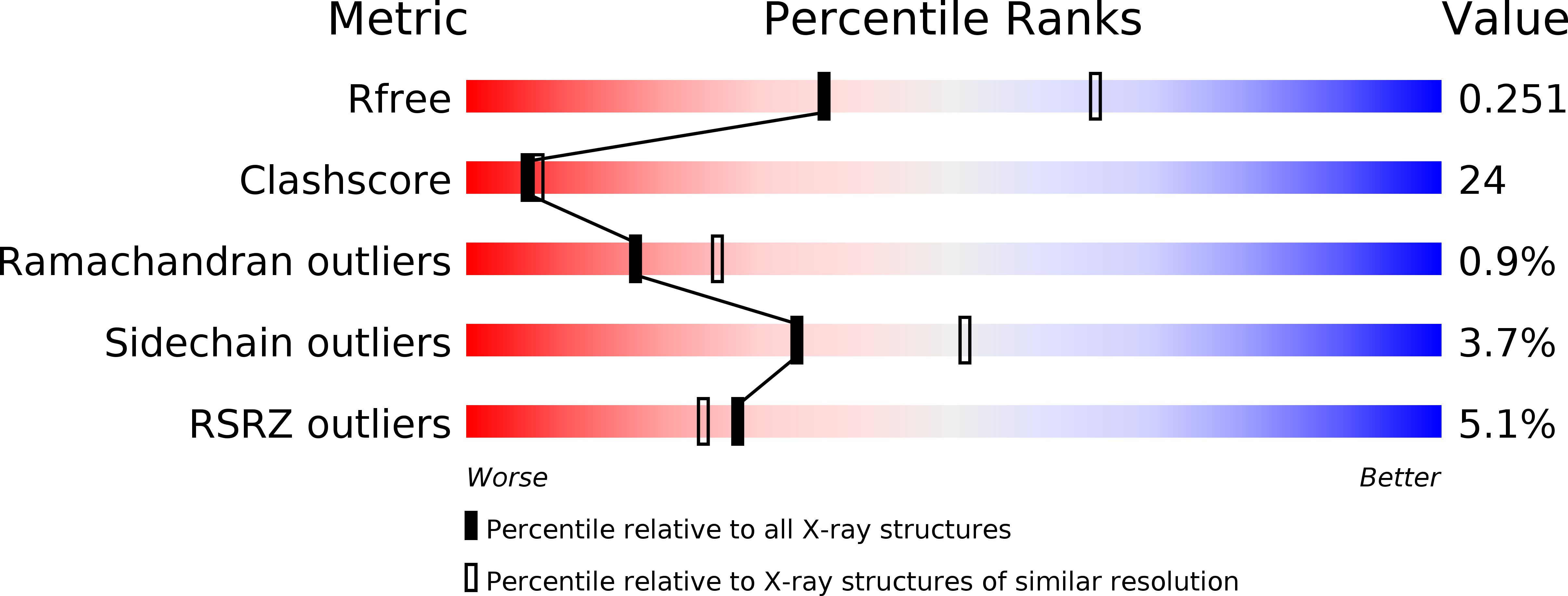
Resolution:
2.63 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.21
Space Group:
C 1 2 1