Deposition Date
2010-04-08
Release Date
2010-04-21
Last Version Date
2025-03-26
Entry Detail
PDB ID:
3MHS
Title:
Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde
Biological Source:
Source Organism:
Saccharomyces cerevisiae (Taxon ID: 4932)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
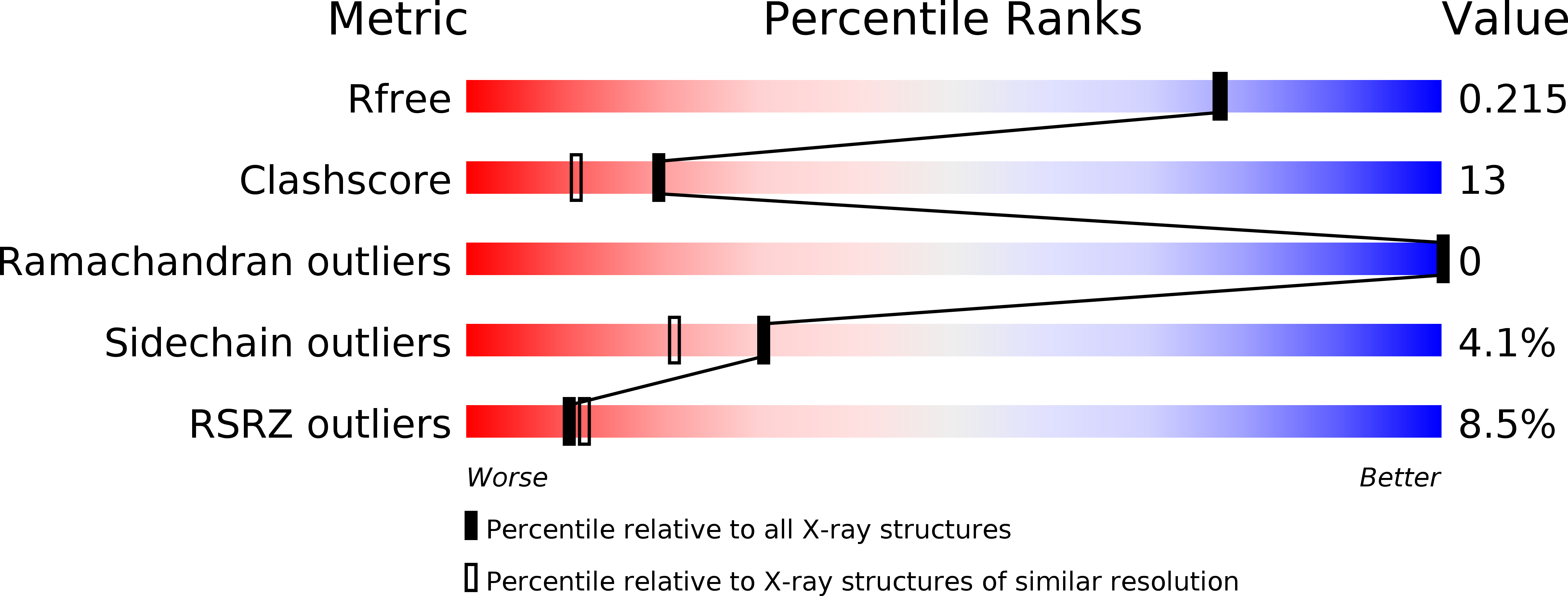
Resolution:
1.89 Å
R-Value Free:
0.20
R-Value Work:
0.15
R-Value Observed:
0.16
Space Group:
P 21 21 21