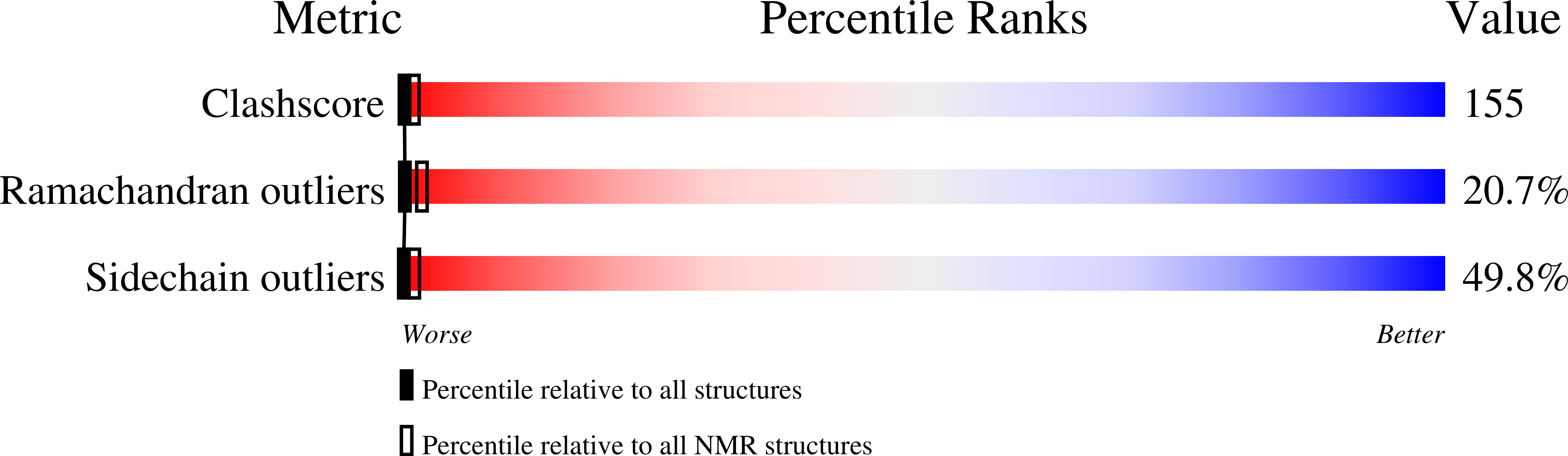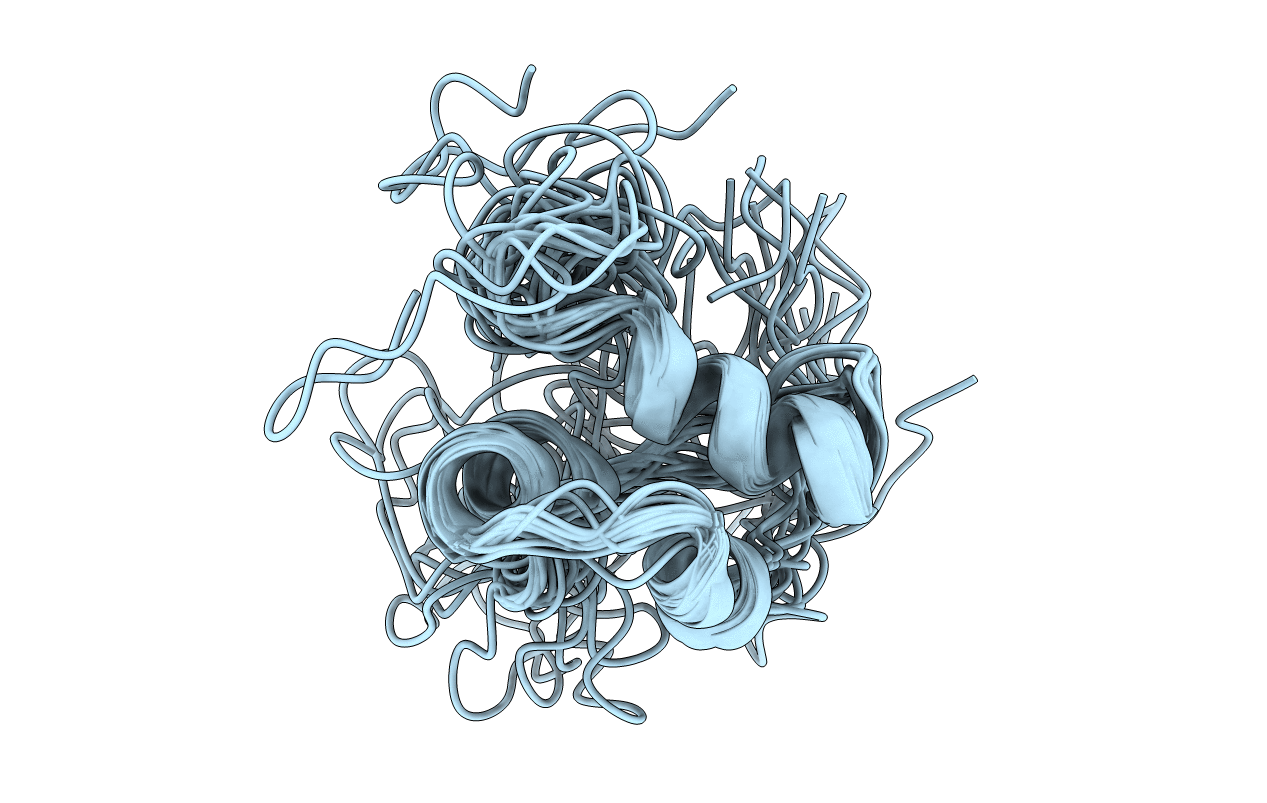
Deposition Date
1999-04-13
Release Date
2000-05-23
Last Version Date
2024-10-30
Entry Detail
PDB ID:
3LRI
Keywords:
Title:
Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
15
Selection Criteria:
LOWEST ENERGY