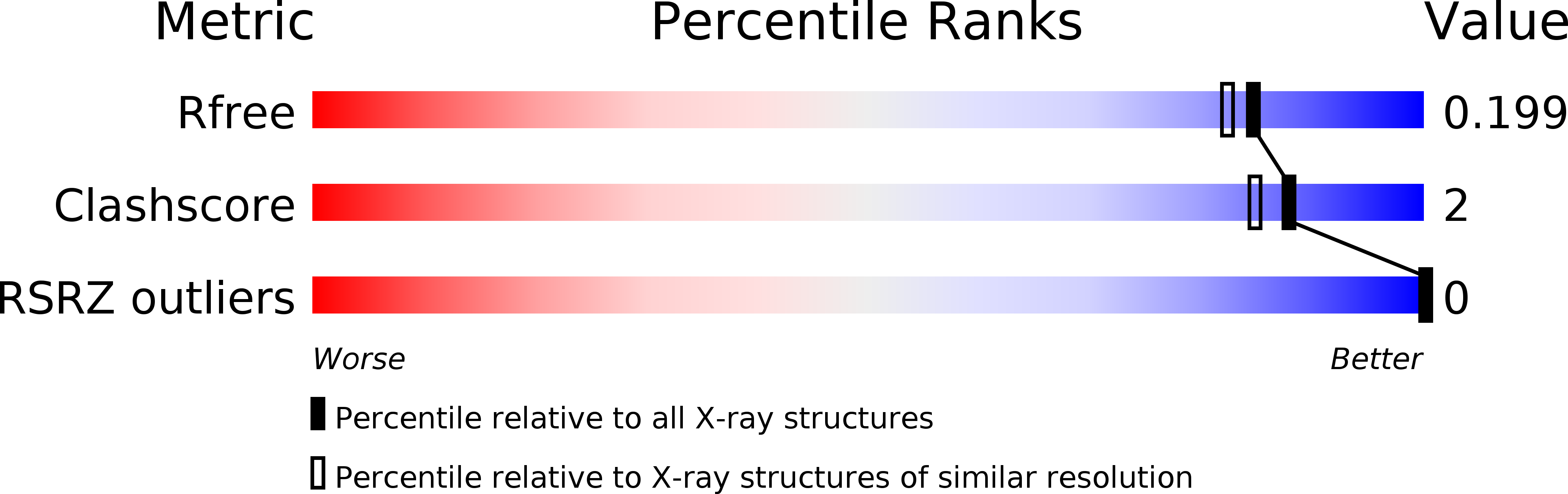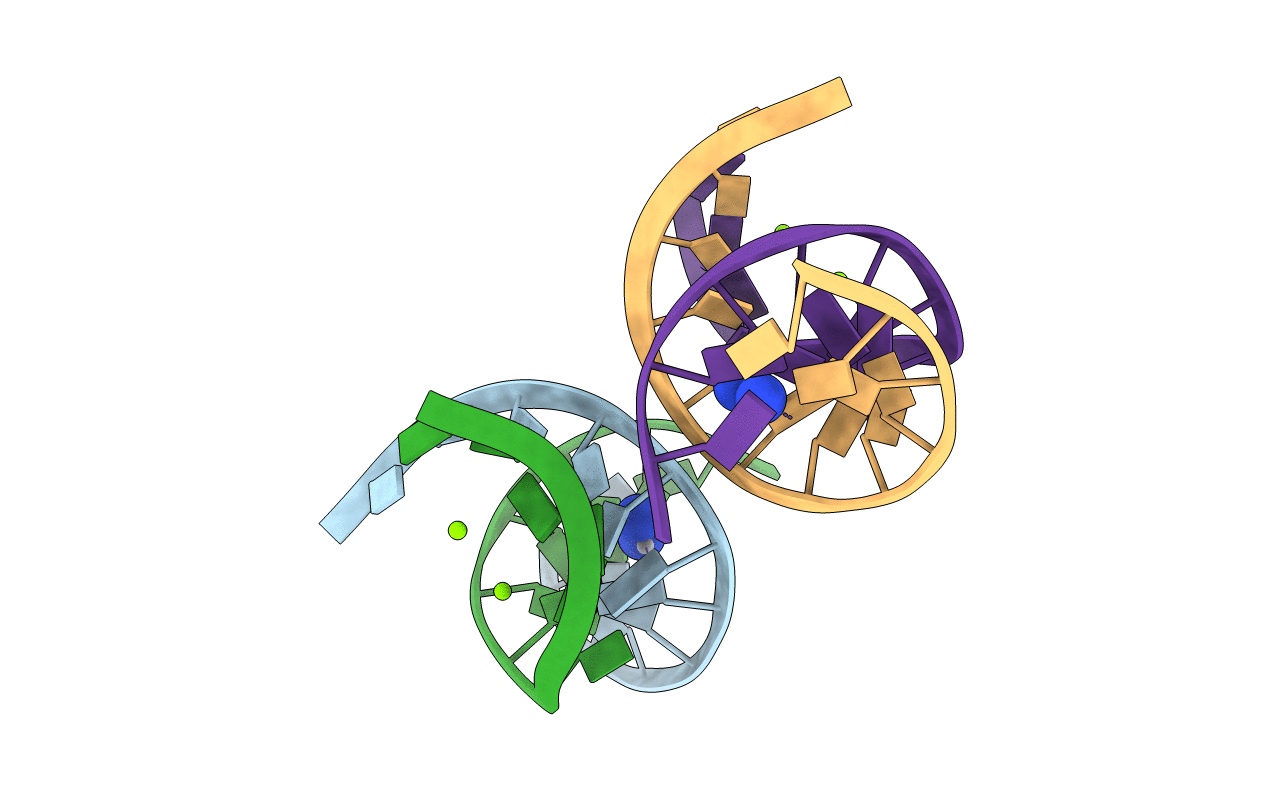
Deposition Date
2010-02-06
Release Date
2010-08-11
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3LPV
Keywords:
Title:
X-ray crystal structure of duplex DNA containing a cisplatin 1,2-d(GpG) intrastrand cross-link
Method Details:
Experimental Method:
Resolution:
1.77 Å
R-Value Free:
0.19
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1