Deposition Date
2010-02-03
Release Date
2010-03-09
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3LNZ
Keywords:
Title:
Crystal structure of human MDM2 with a 12-mer peptide inhibitor PMI (N8A mutant)
Method Details:
Experimental Method:
Resolution:
1.95 Å
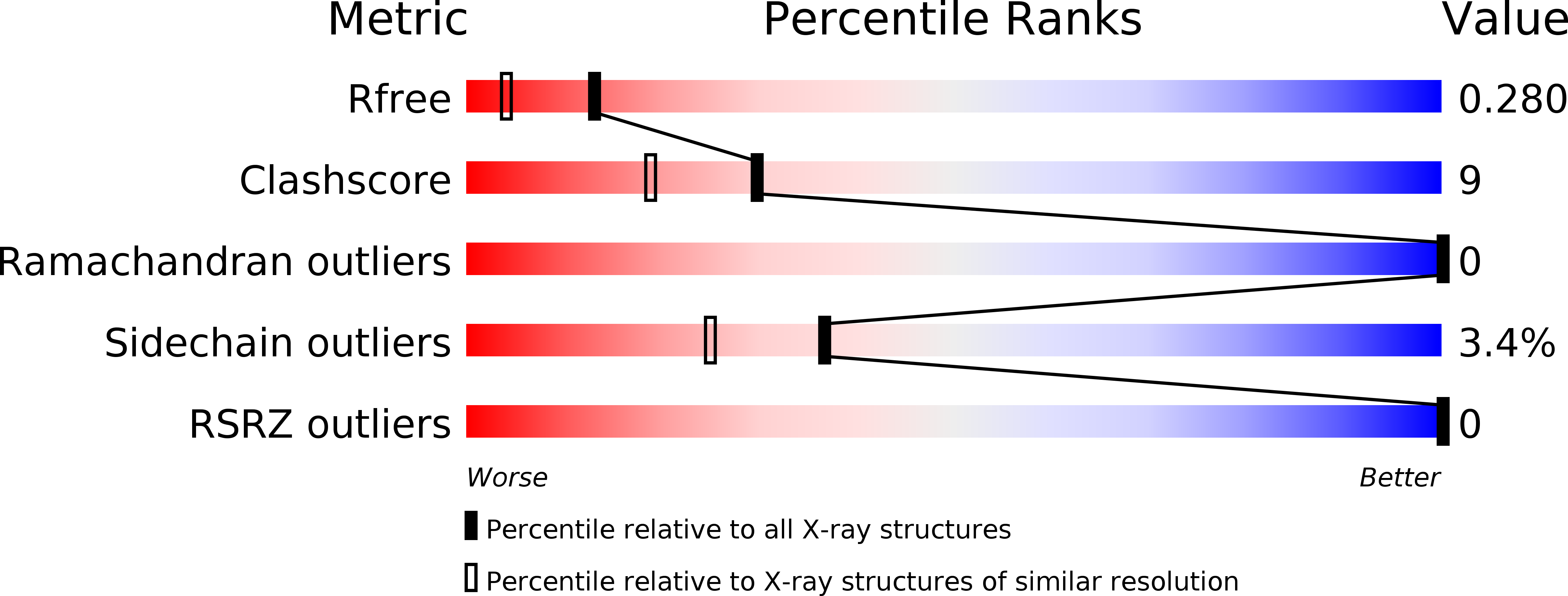
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.22
Space Group:
P 32 1 2