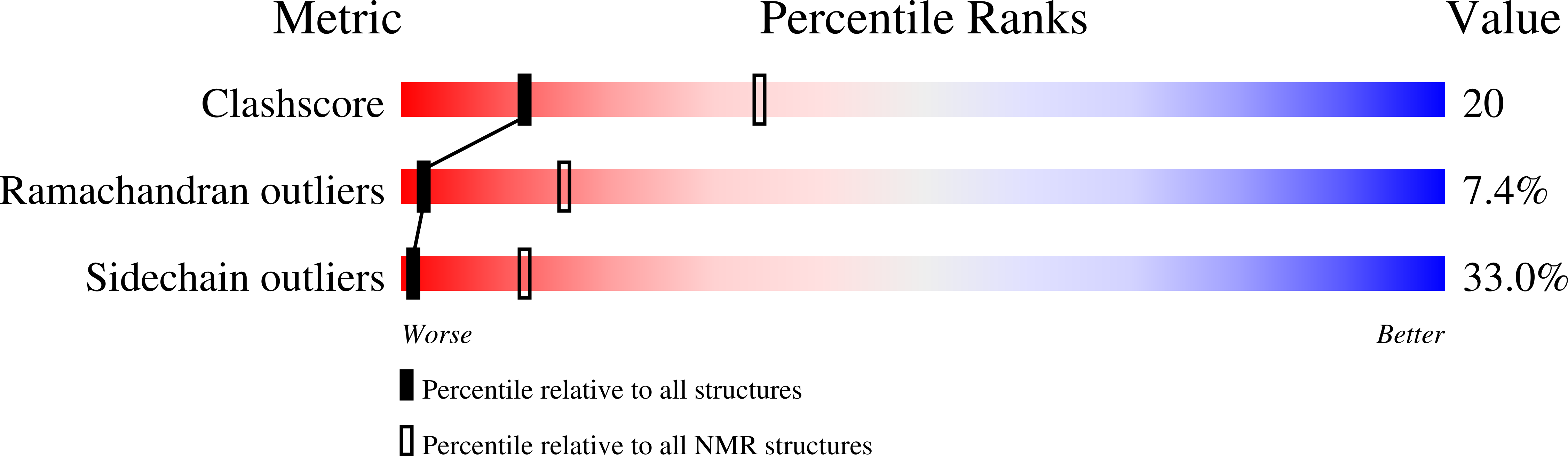
Deposition Date
1997-05-20
Release Date
1997-11-26
Last Version Date
2024-10-23
Entry Detail
PDB ID:
3LEU
Keywords:
Title:
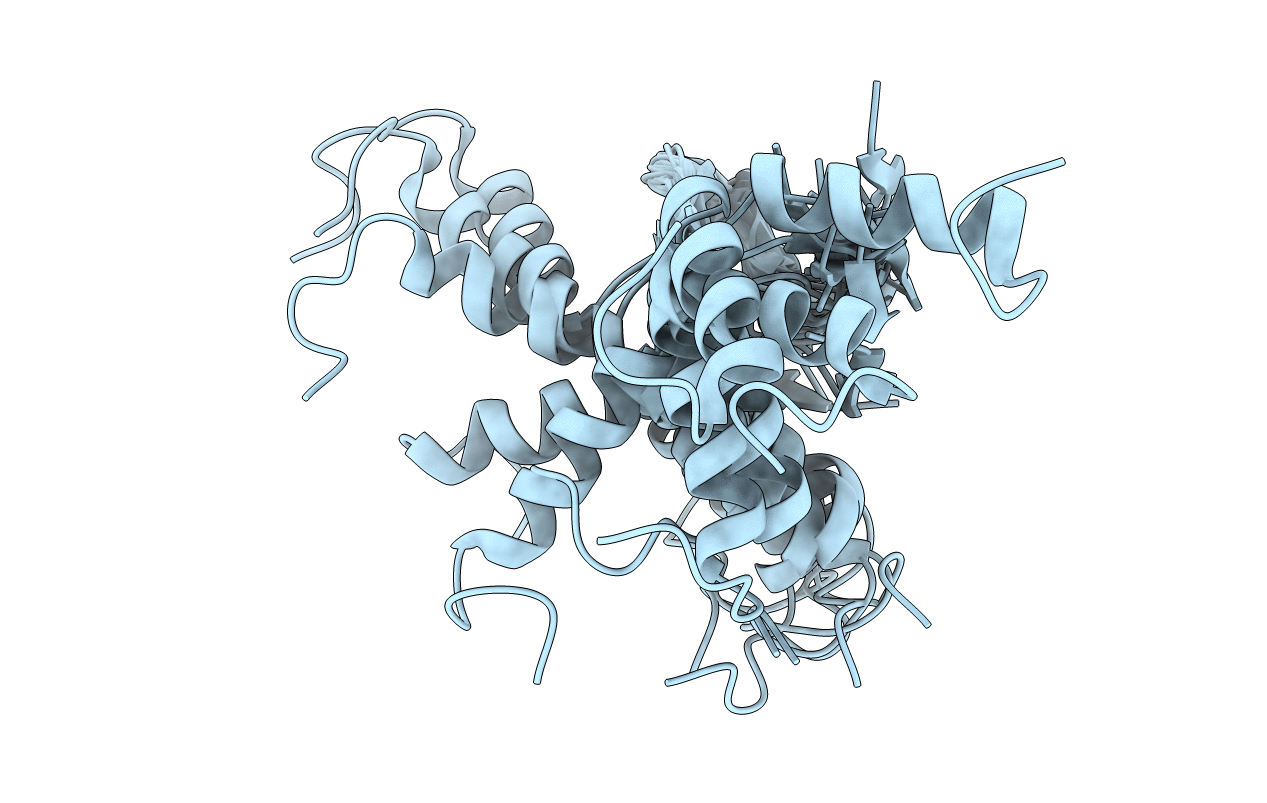
HIGH RESOLUTION 1H NMR STUDY OF LEUCOCIN A IN DODECYLPHOSPHOCHOLINE MICELLES, 19 STRUCTURES (1:40 RATIO OF LEUCOCIN A:DPC) (0.1% TFA)
Biological Source:
Source Organism(s):
Leuconostoc gelidum (Taxon ID: 1244)
Method Details:
Experimental Method:
Conformers Calculated:
25
Conformers Submitted:
19
Selection Criteria:
LEAST RESTRAINT VIOLATION