Deposition Date
2010-01-12
Release Date
2010-08-11
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3LDD
Keywords:
Title:
High resolution open MthK pore structure crystallized in 100 mM K+ and further soaked in 99 mM Na+/1 mM K+.
Biological Source:
Source Organism(s):
Methanothermobacter thermautotrophicus (Taxon ID: 187420)
Expression System(s):
Method Details:
Experimental Method:
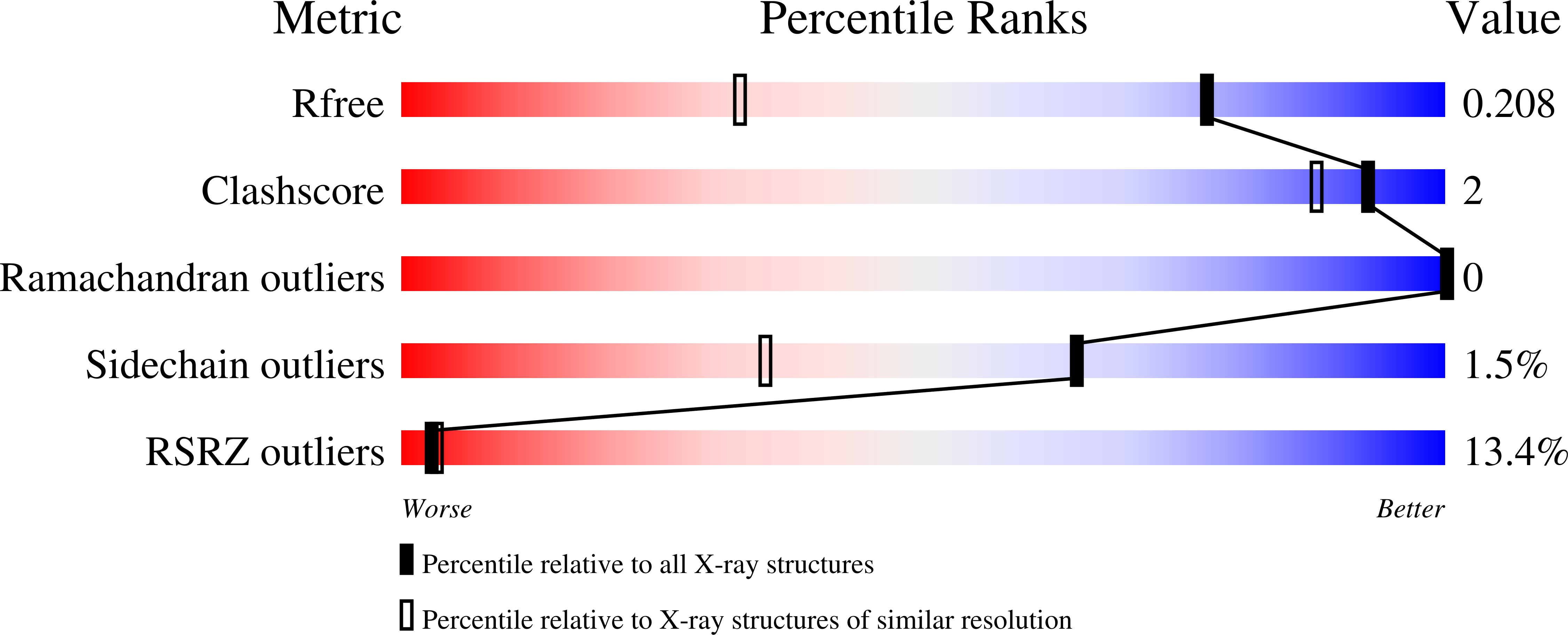
Resolution:
1.45 Å
R-Value Free:
0.21
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 4 21 2