Deposition Date
2010-01-05
Release Date
2010-03-02
Last Version Date
2024-03-20
Entry Detail
PDB ID:
3L9H
Keywords:
Title:
X-ray structure of mitotic kinesin-5 (KSP, KIF11, Eg5)in complex with the hexahydro-2H-pyrano[3,2-c]quinoline EMD 534085
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
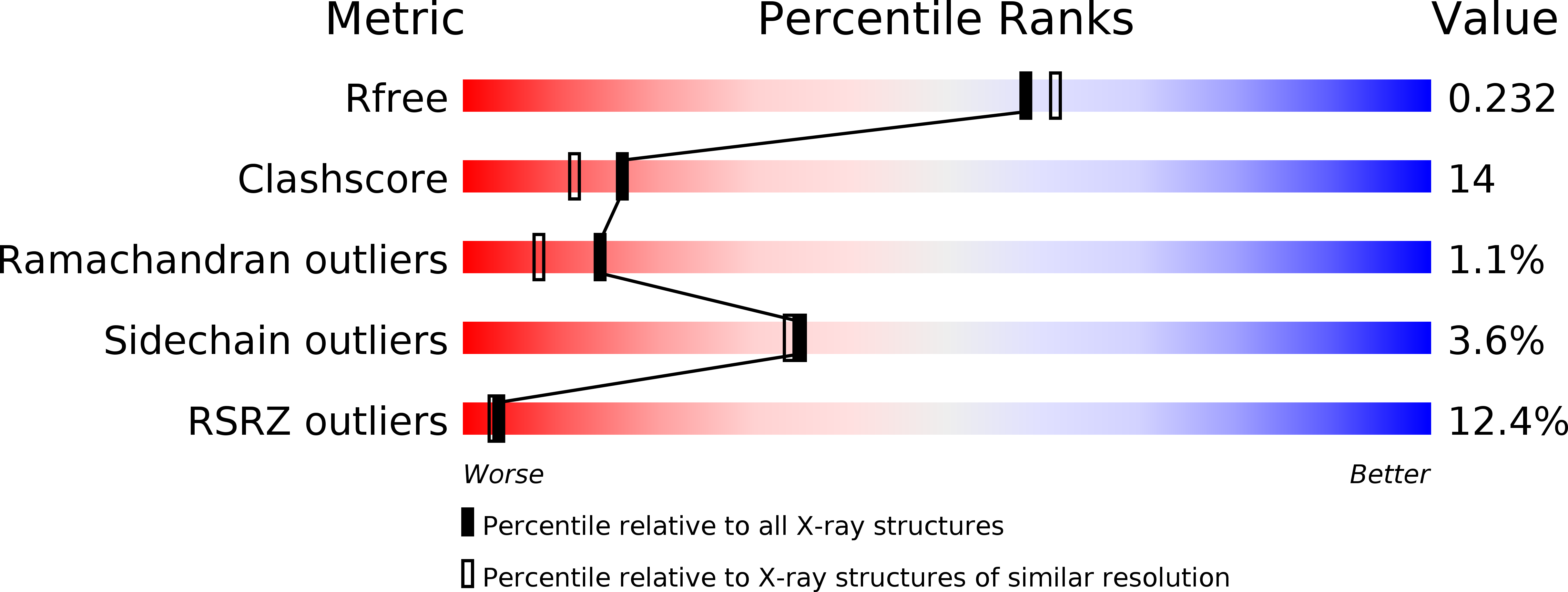
Resolution:
2.00 Å
R-Value Free:
0.25
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
C 1 2 1