Deposition Date
2009-10-29
Release Date
2010-08-18
Last Version Date
2024-10-30
Entry Detail
PDB ID:
3KH2
Keywords:
Title:
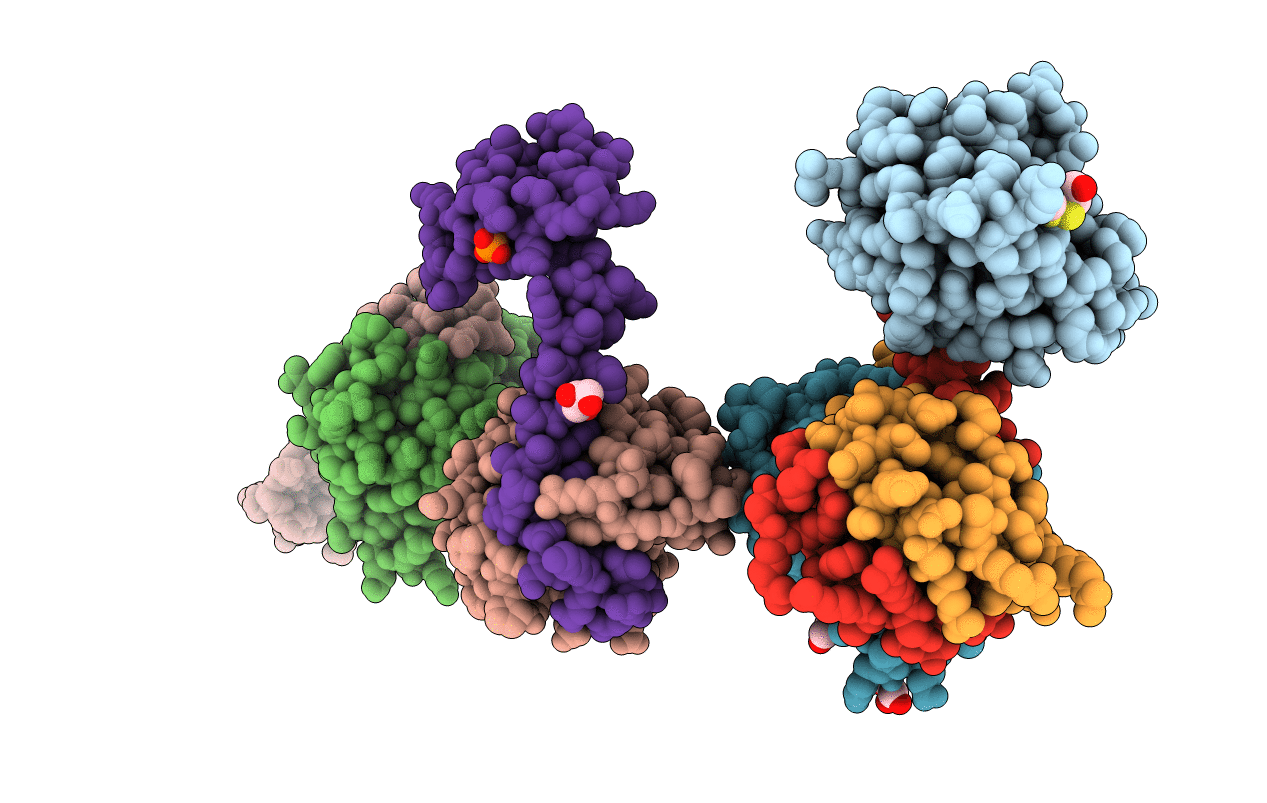
Crystal structure of the P1 bacteriophage Doc toxin (F68S) in complex with the Phd antitoxin (L17M/V39A). Northeast Structural Genomics targets ER385-ER386
Biological Source:
Source Organism(s):
Bacteriophage P1 (Taxon ID: 10678)
Expression System(s):
Method Details:
Experimental Method:
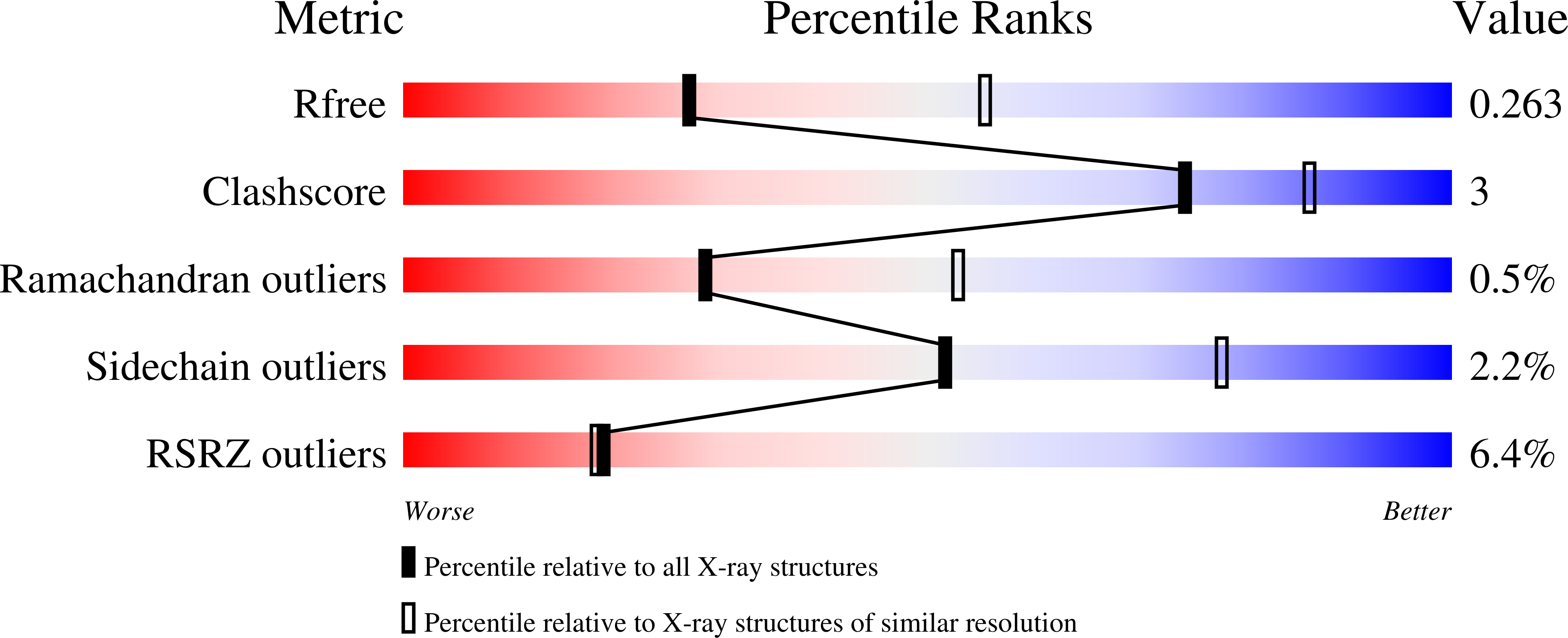
Resolution:
2.71 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21