Deposition Date
2009-10-28
Release Date
2009-12-08
Last Version Date
2024-10-30
Entry Detail
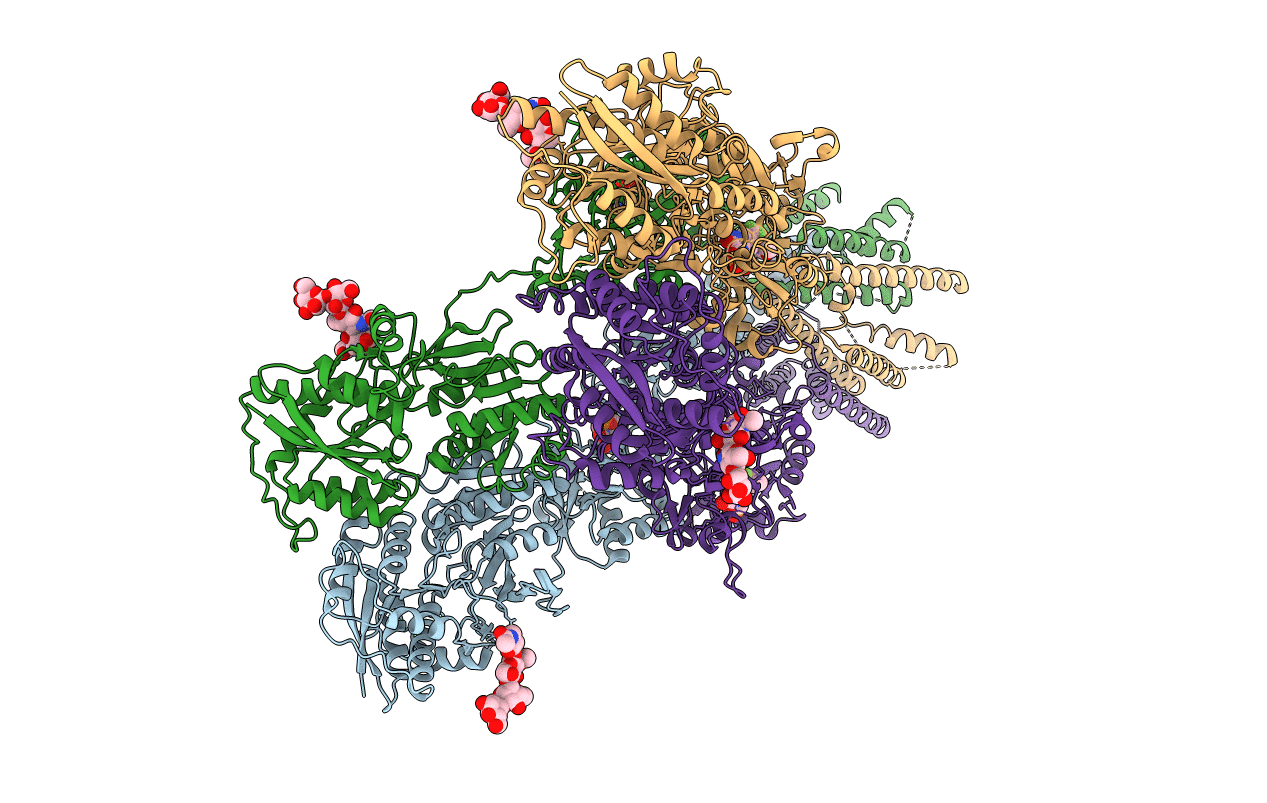
PDB ID:
3KG2
Keywords:
Title:
AMPA subtype ionotropic glutamate receptor in complex with competitive antagonist ZK 200775
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.60 Å
R-Value Free:
0.29
R-Value Work:
0.28
R-Value Observed:
0.28
Space Group:
P 1


