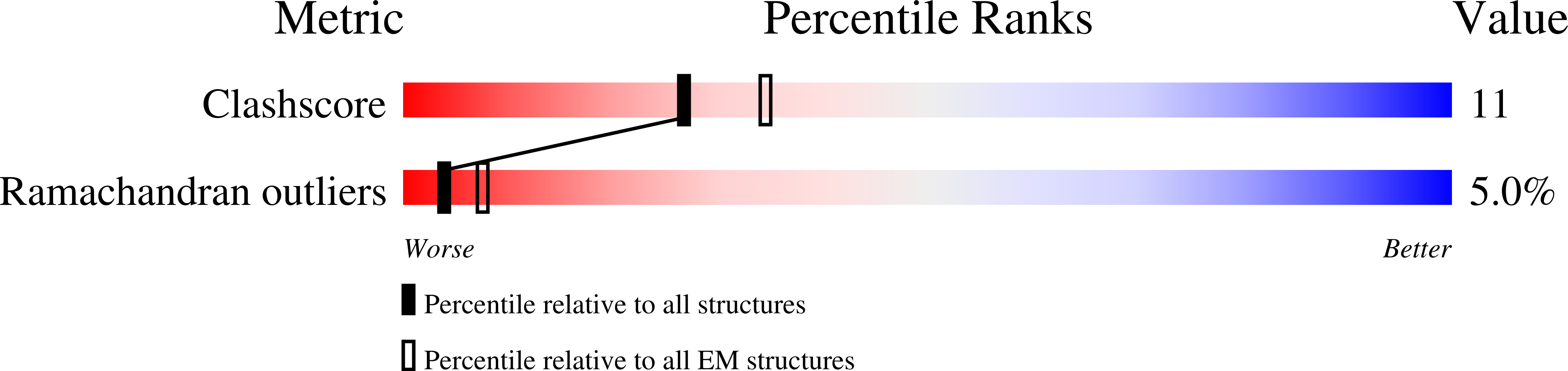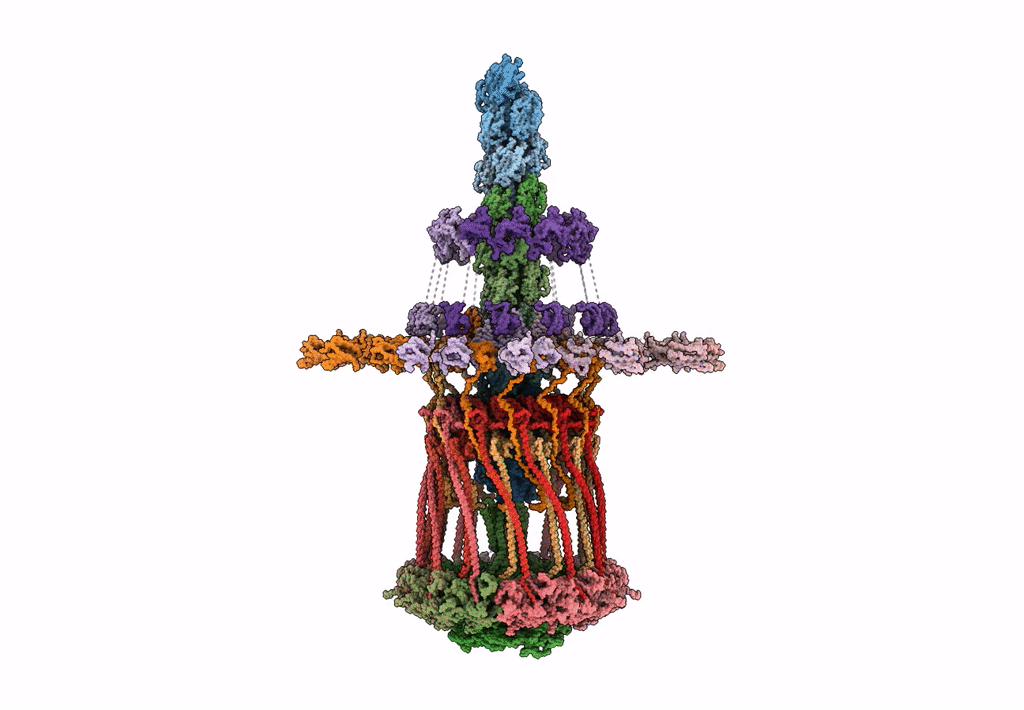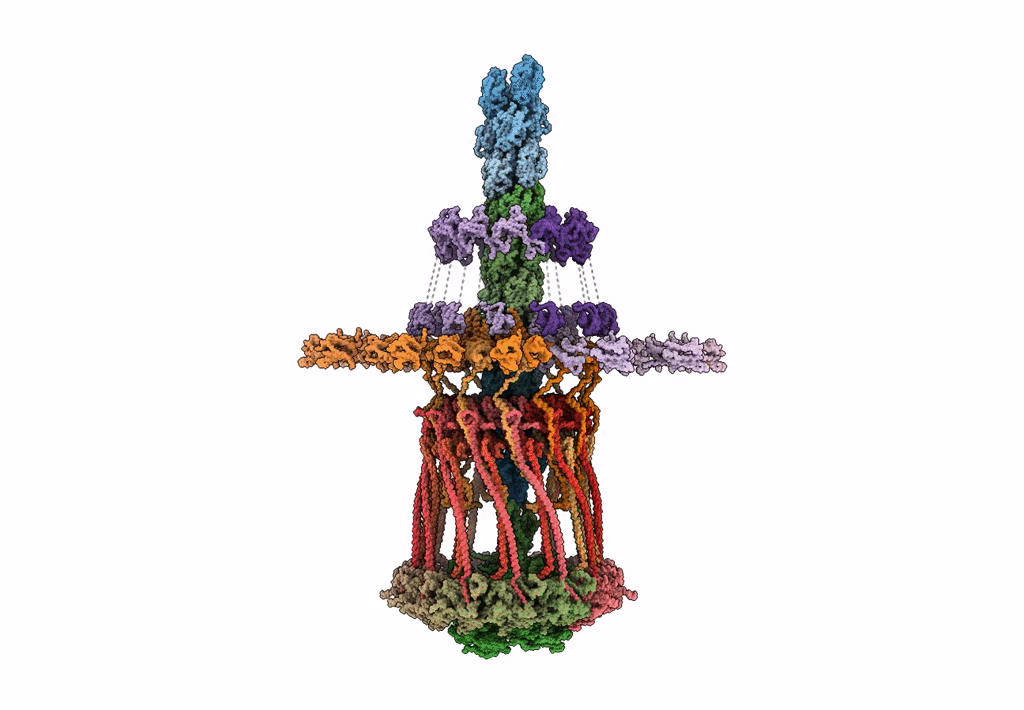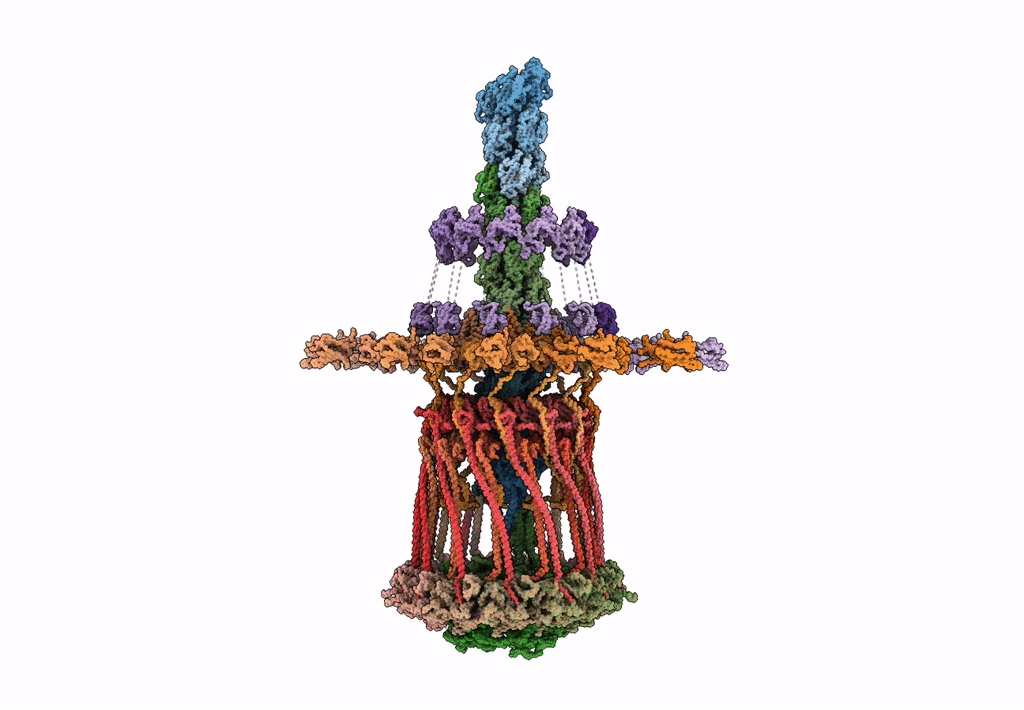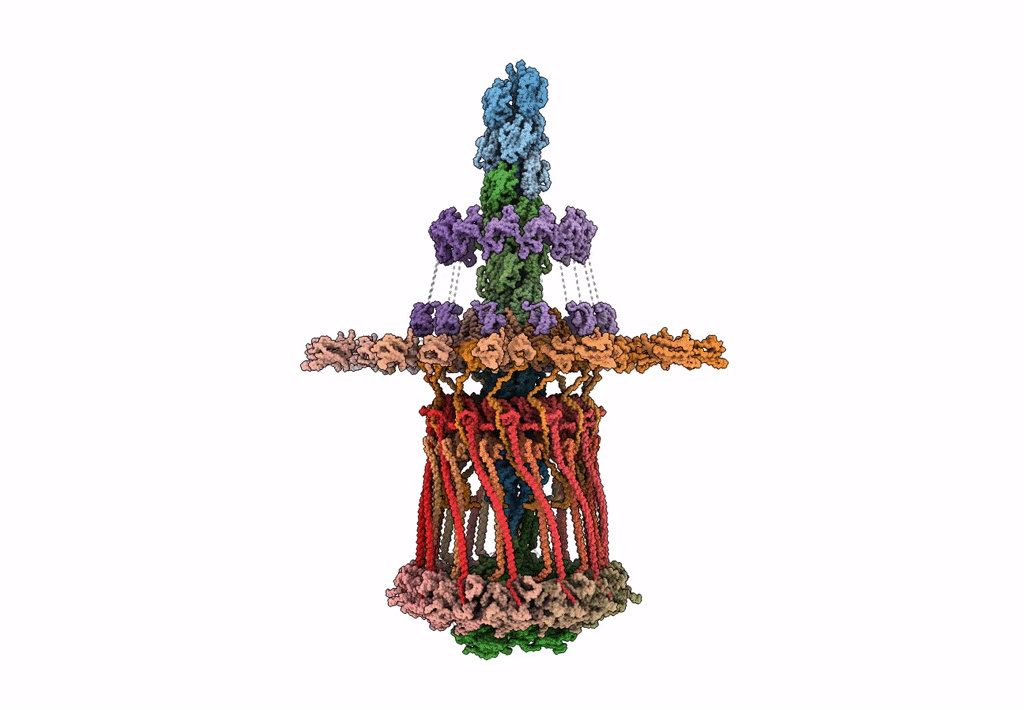
Deposition Date
2015-11-24
Release Date
2016-03-16
Last Version Date
2024-11-06
Entry Detail
PDB ID:
3JC8
Keywords:
Title:
Architectural model of the type IVa pilus machine in a piliated state
Biological Source:
Source Organism(s):
Myxococcus xanthus DK 1622 (Taxon ID: 246197)
Method Details:
Experimental Method:
Aggregation State:
PARTICLE
Reconstruction Method:
TOMOGRAPHY