Deposition Date
2015-01-16
Release Date
2015-03-11
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3J9G
Keywords:
Title:
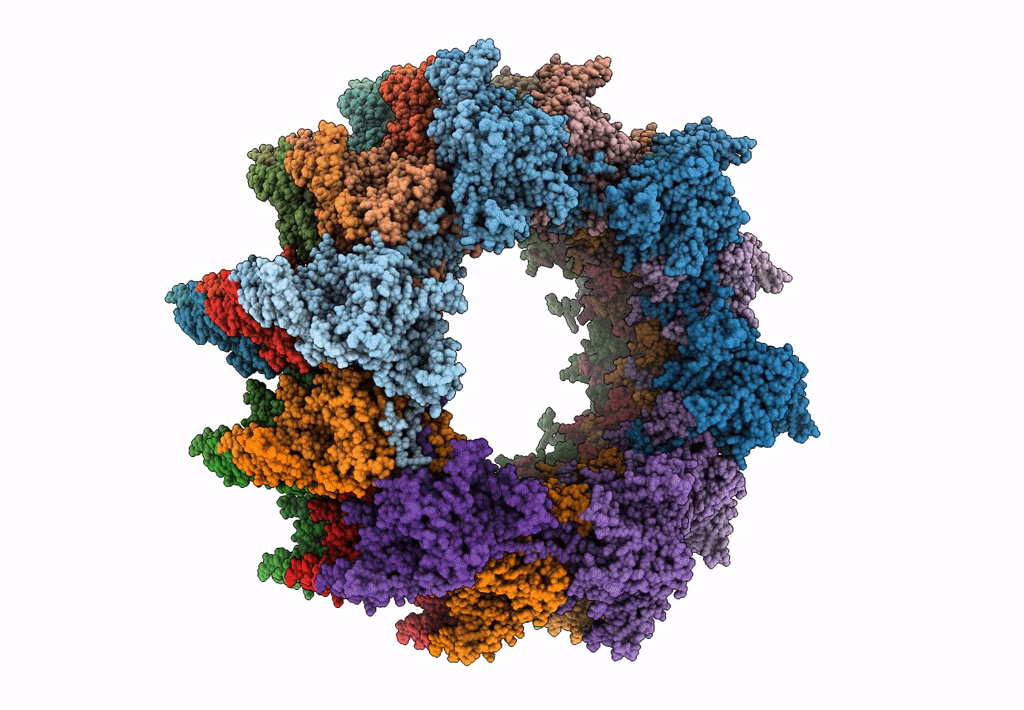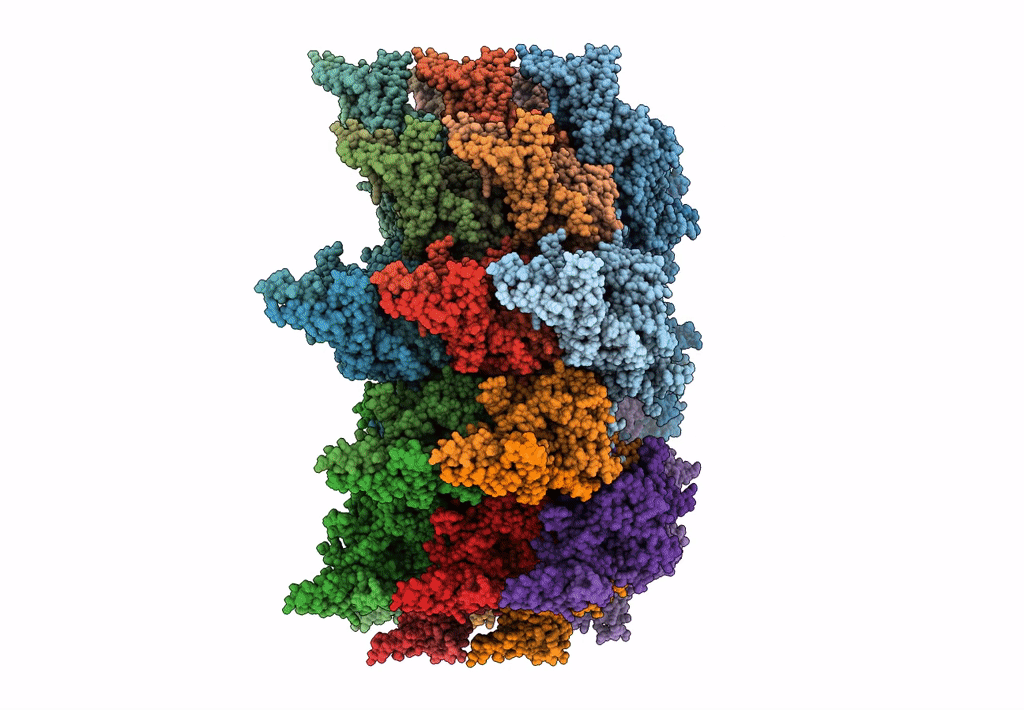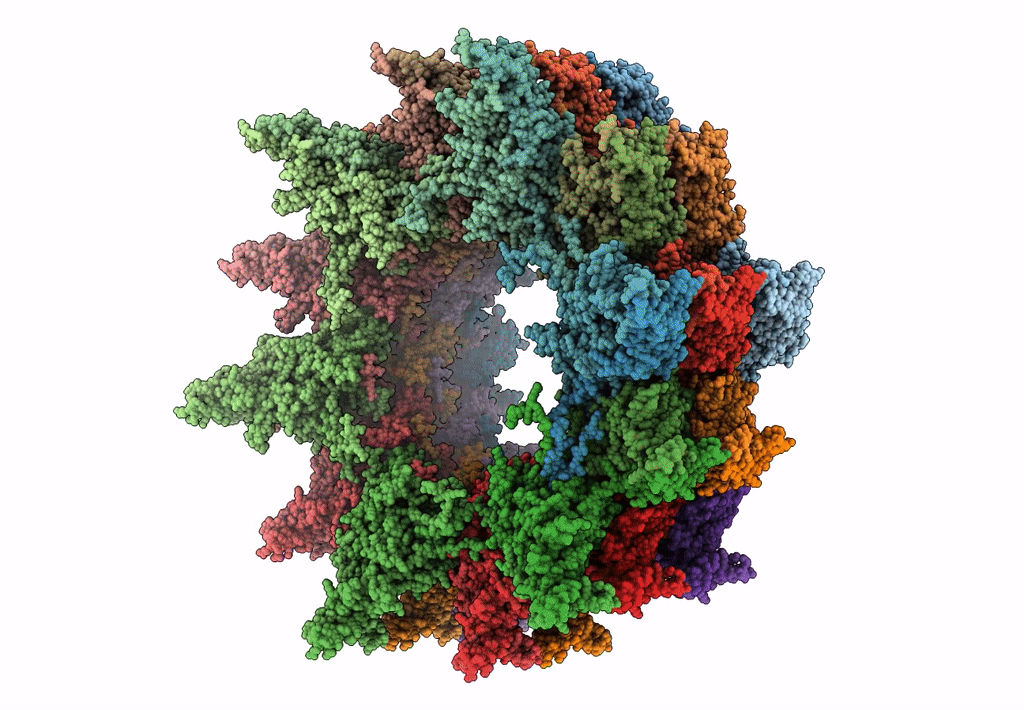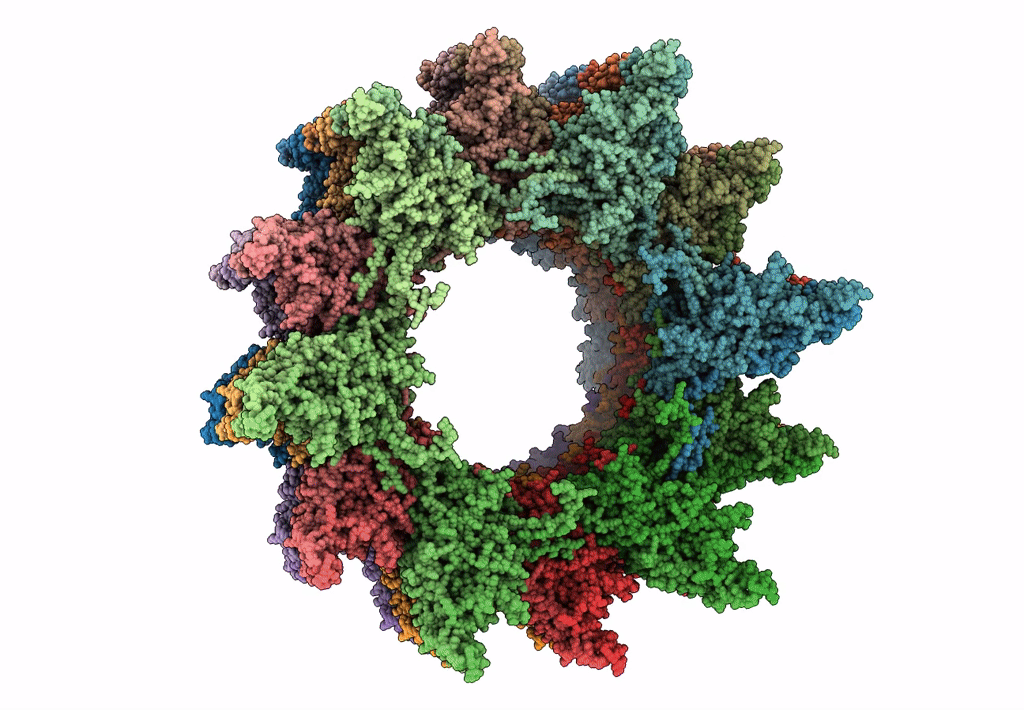
Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM
Biological Source:
Source Organism(s):
Vibrio cholerae O1 biovar El Tor str. N16961 (Taxon ID: 243277)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
FILAMENT
Reconstruction Method:
HELICAL


