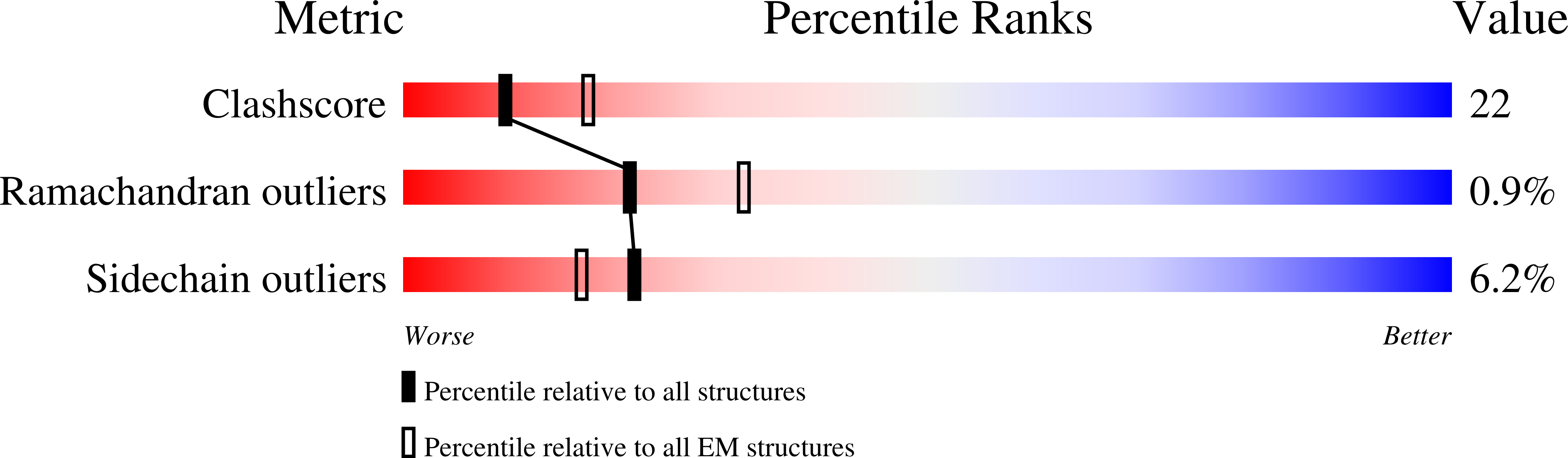
Deposition Date
2015-01-15
Release Date
2015-02-11
Last Version Date
2022-12-21
Entry Detail
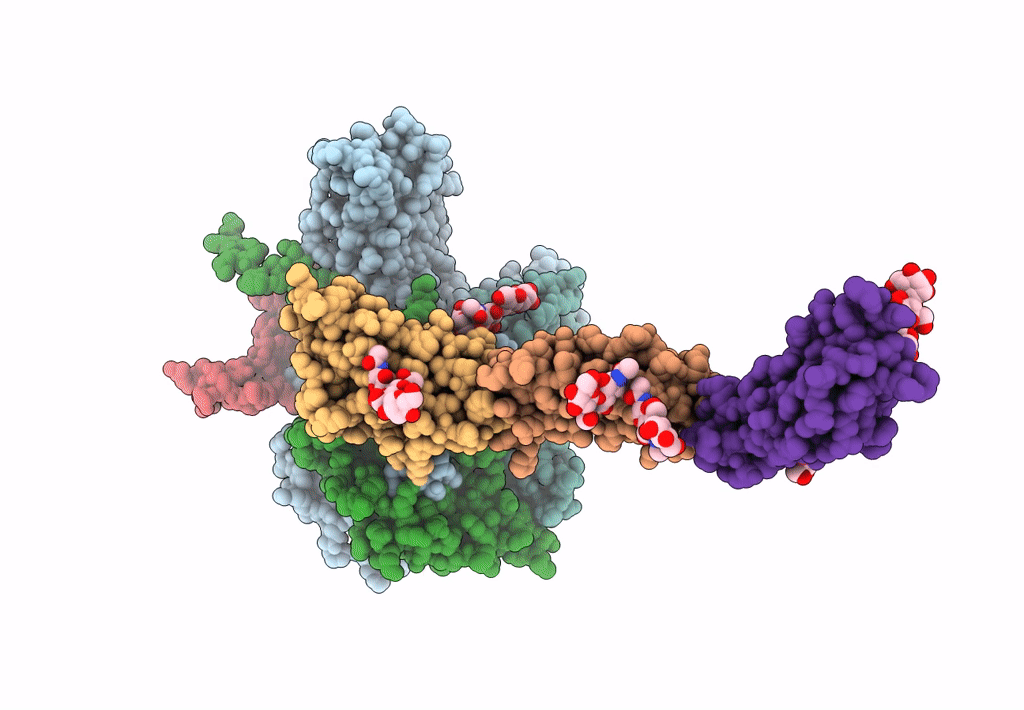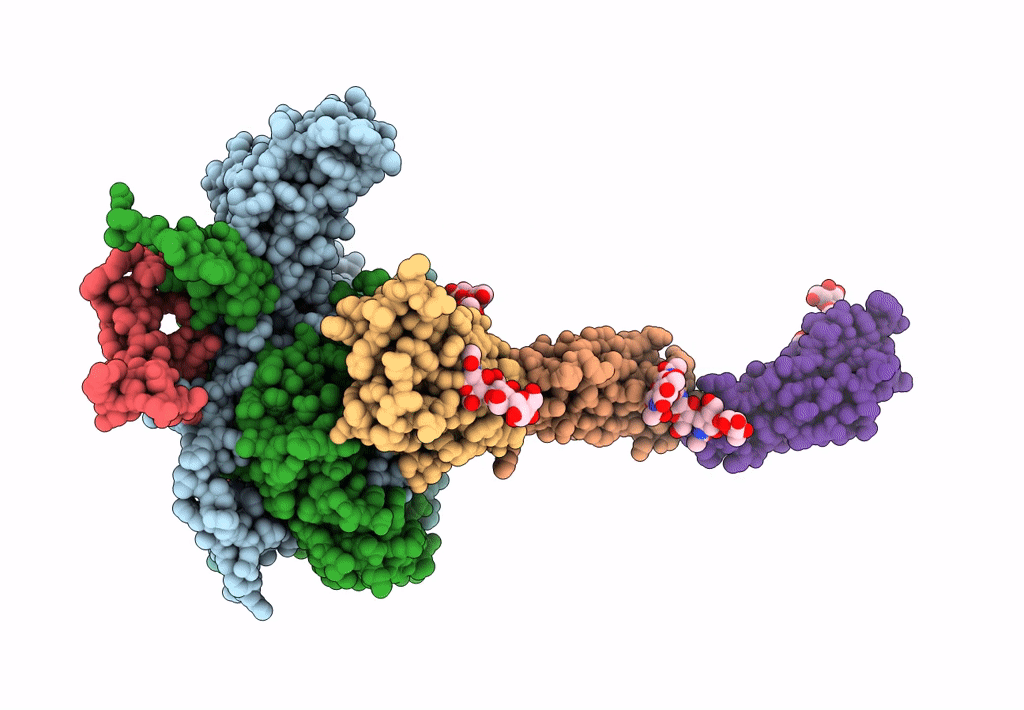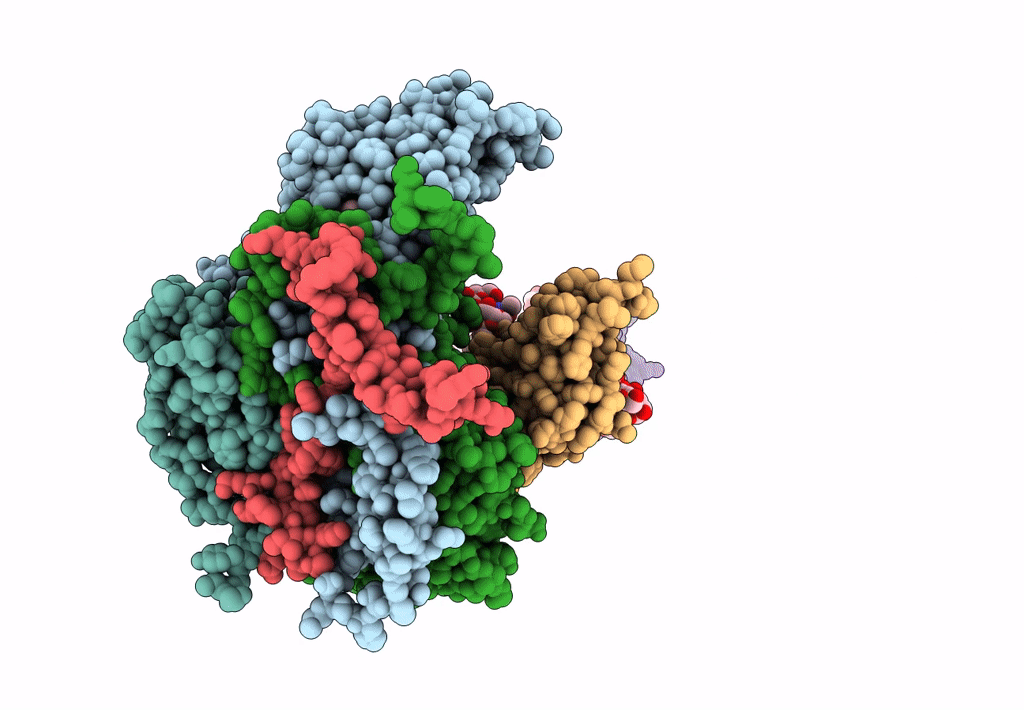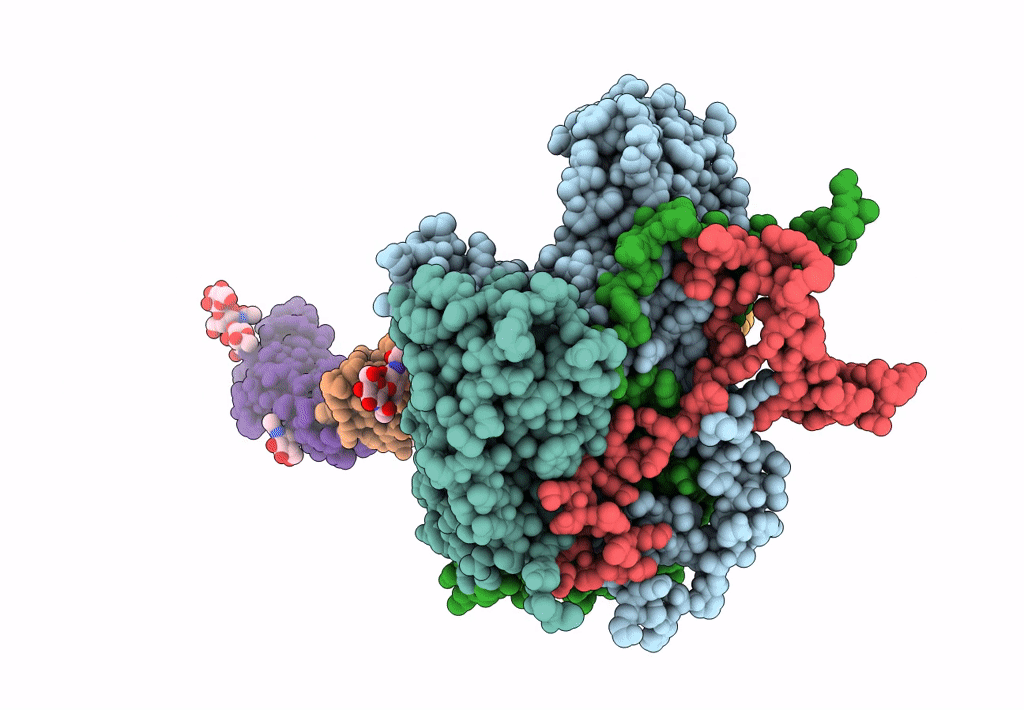
PDB ID:
3J9F
Keywords:
Title:
Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C
Biological Source:
Source Organism(s):
Human poliovirus 1 Mahoney (Taxon ID: 12081)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
Resolution:
9.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE