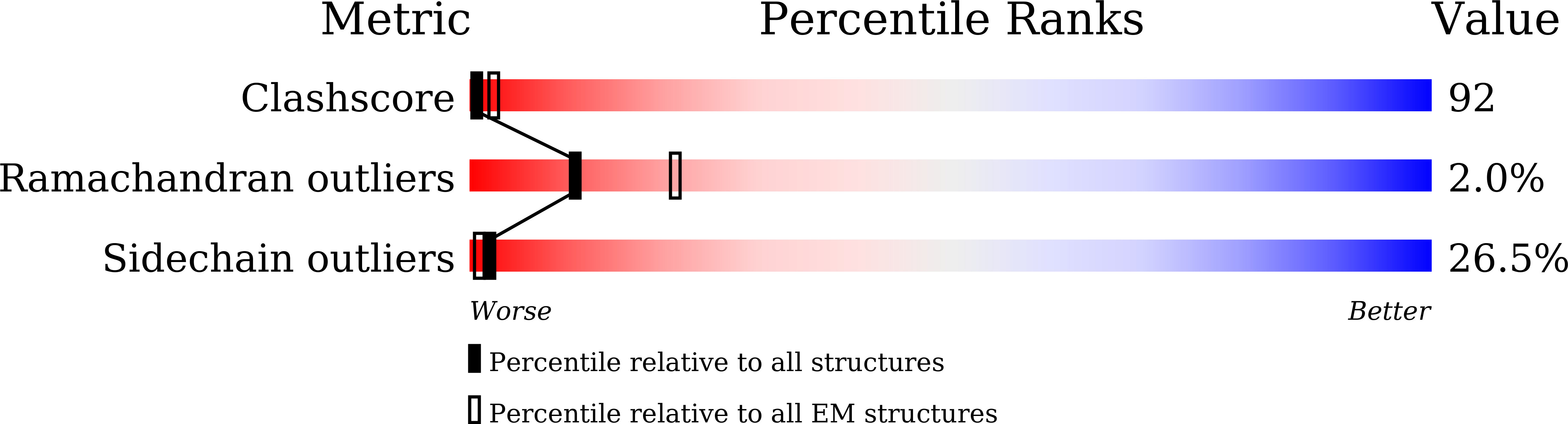
Deposition Date
2014-03-20
Release Date
2014-12-31
Last Version Date
2024-03-20
Entry Detail
PDB ID:
3J6P
Keywords:
Title:
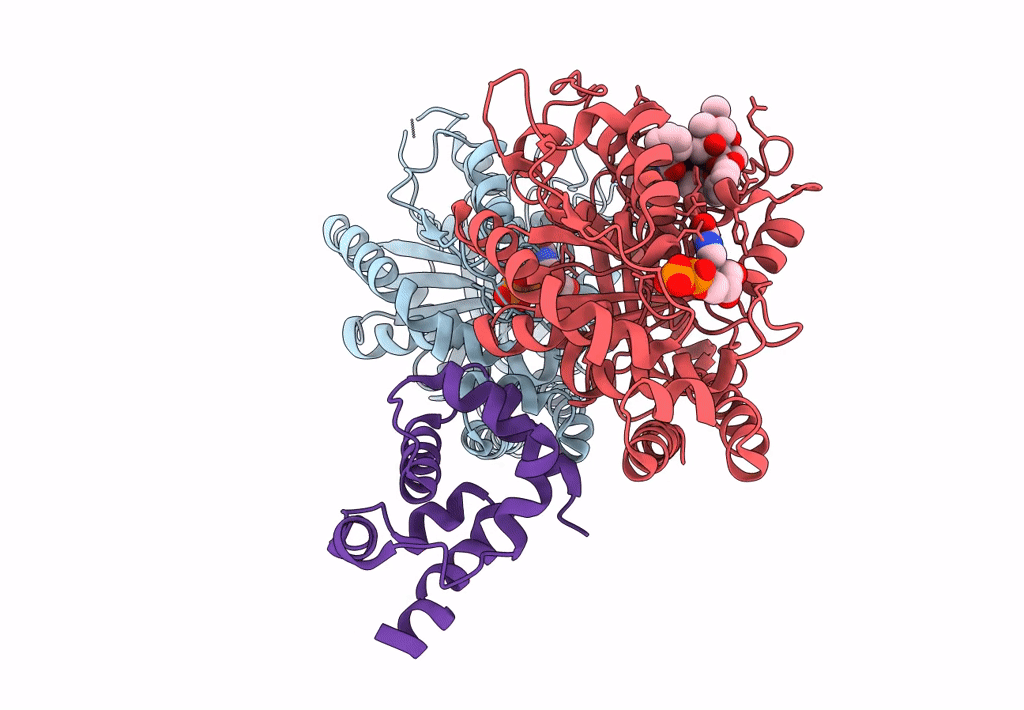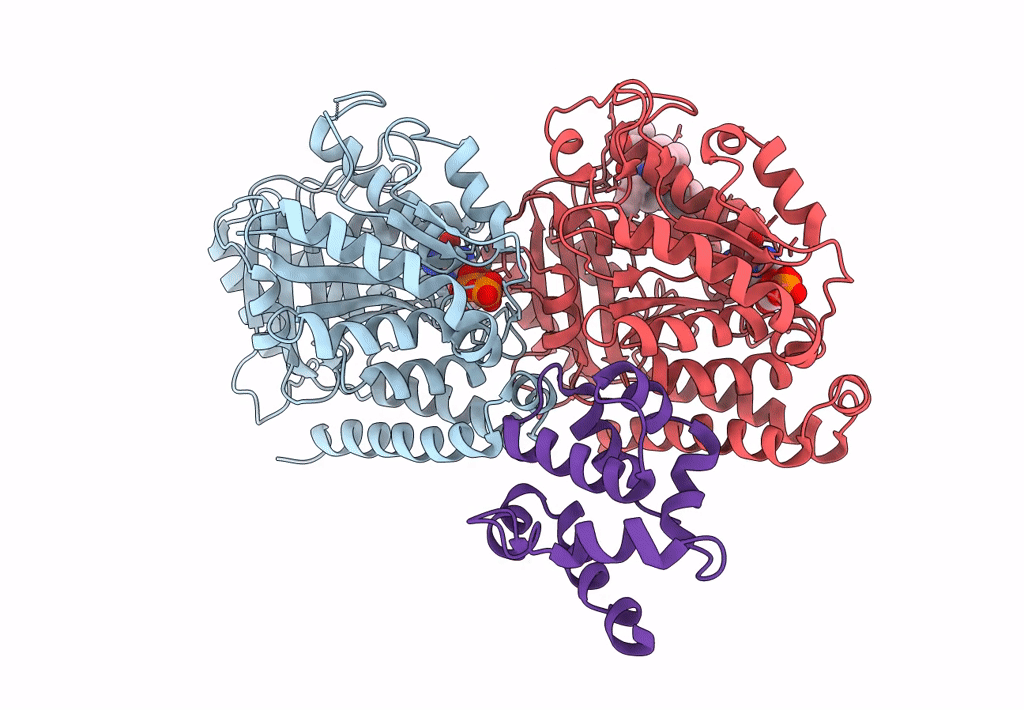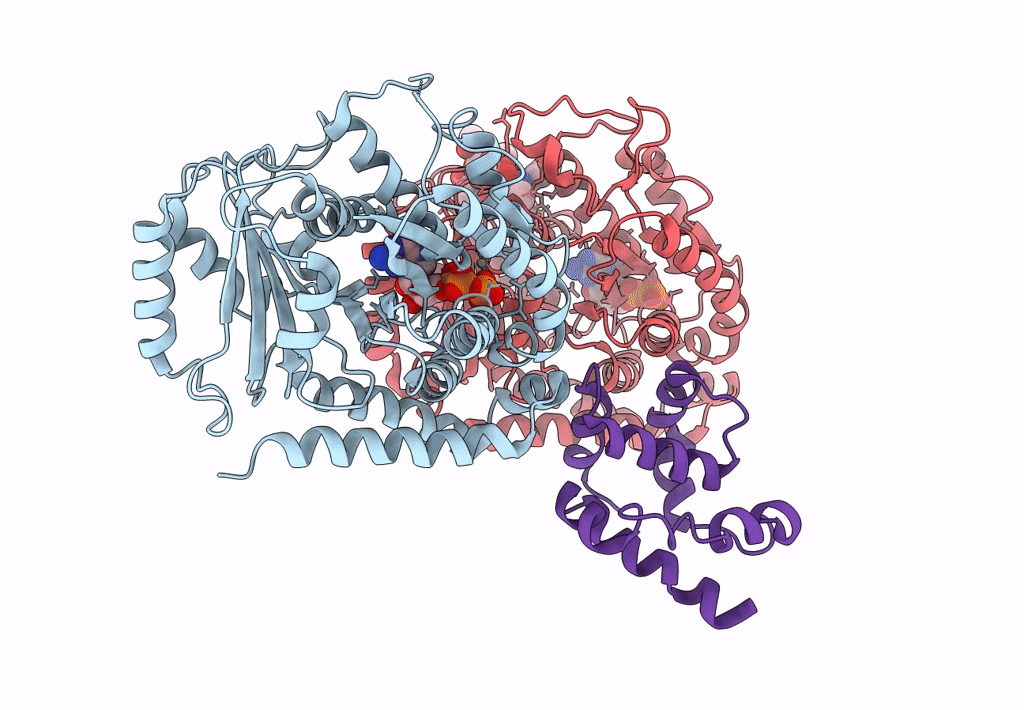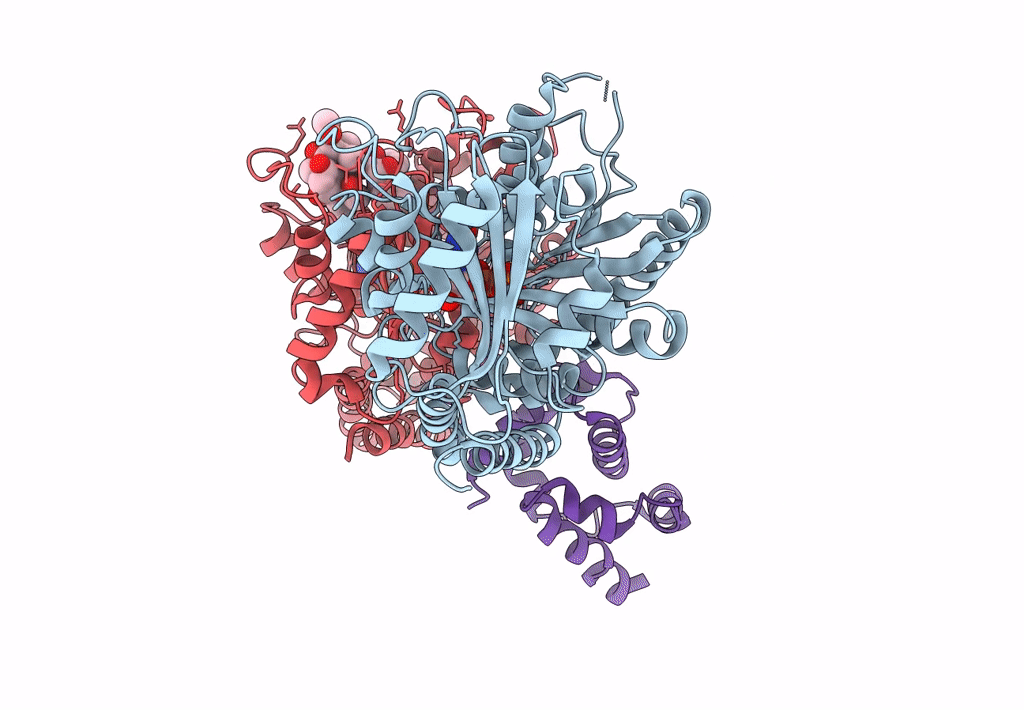
Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map
Biological Source:
Source Organism(s):
Dictyostelium discoideum (Taxon ID: 44689)
Sus scrofa (Taxon ID: 9823)
Sus scrofa (Taxon ID: 9823)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
8.20 Å
Aggregation State:
FILAMENT
Reconstruction Method:
HELICAL