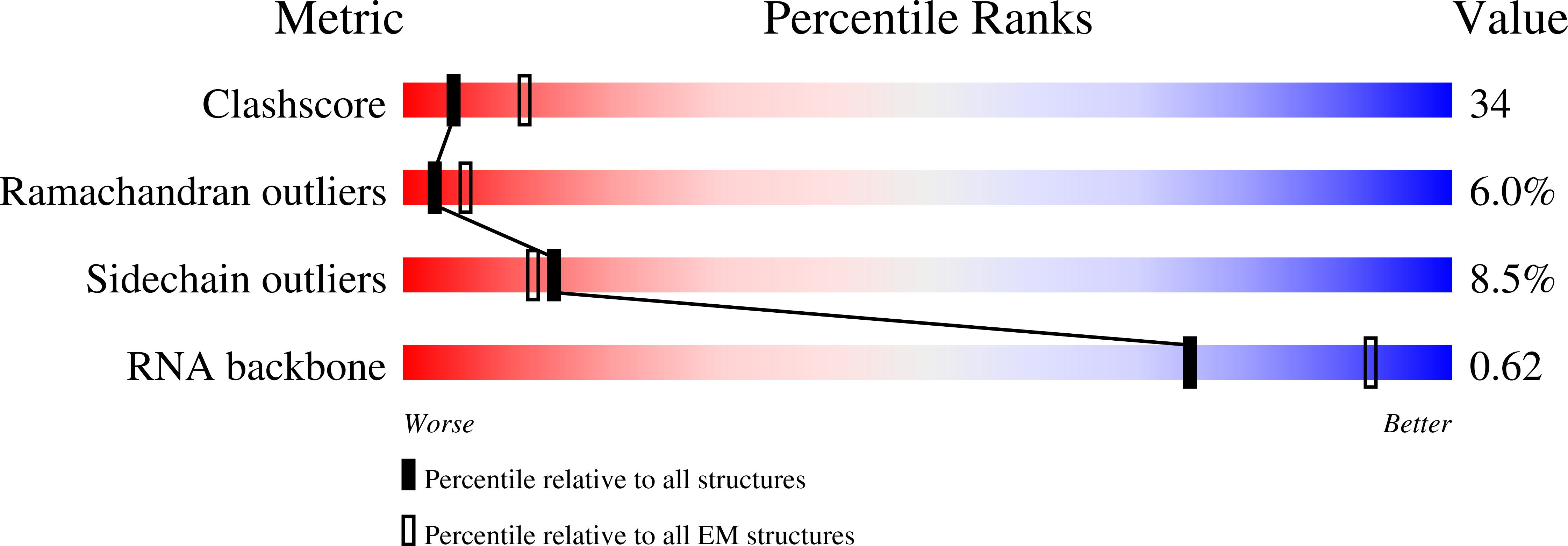
Deposition Date
2011-06-29
Release Date
2012-04-25
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3J0E
Keywords:
Title:
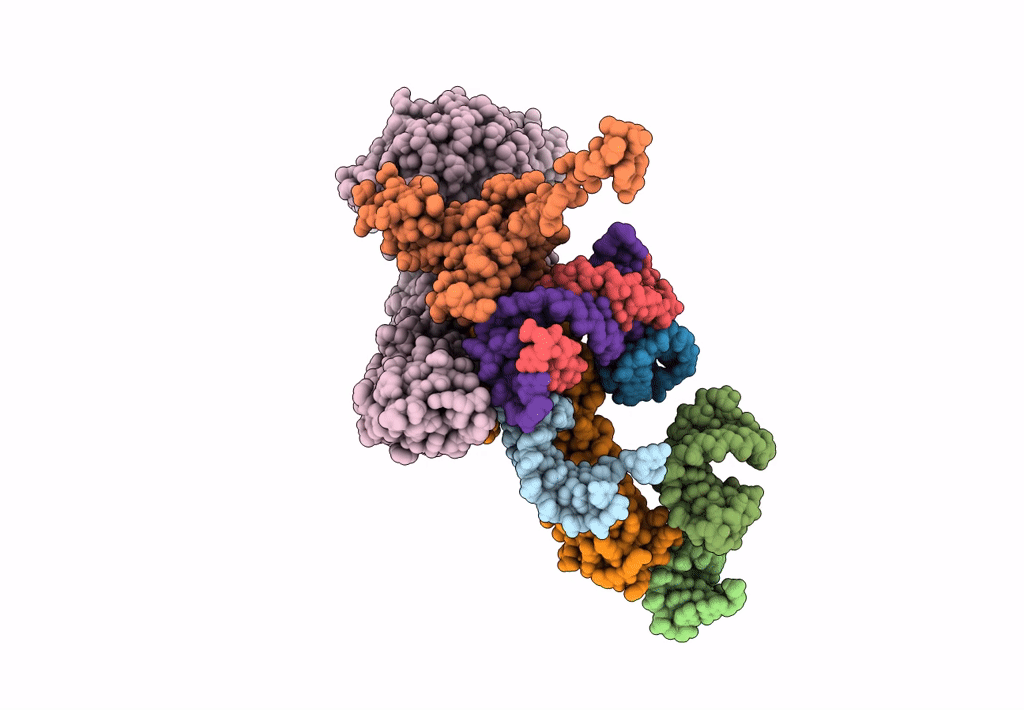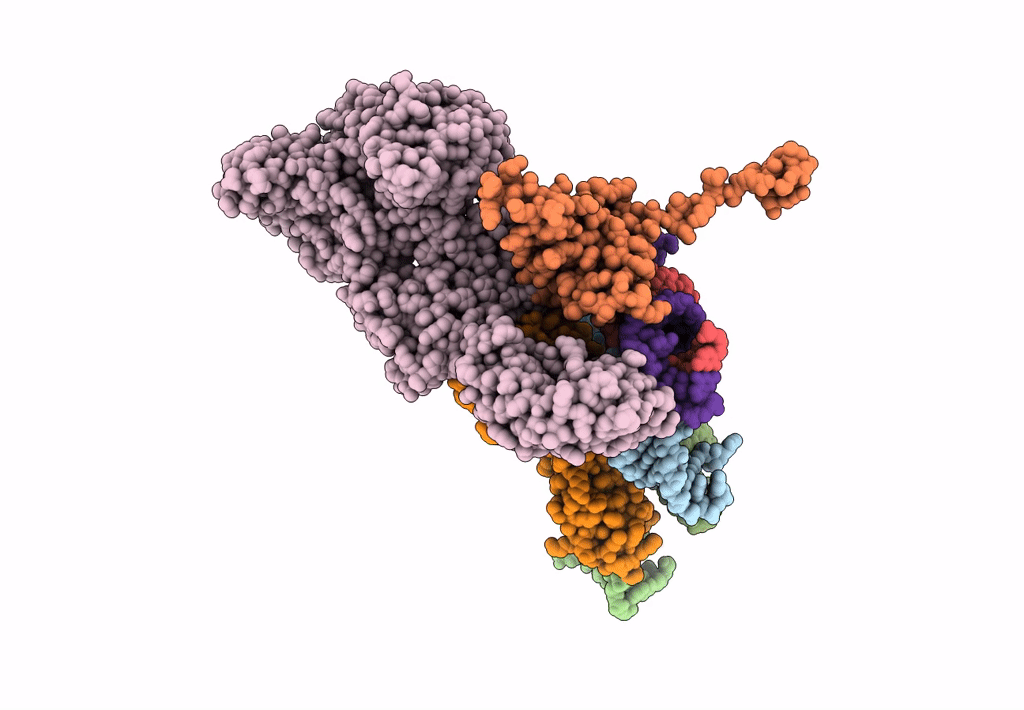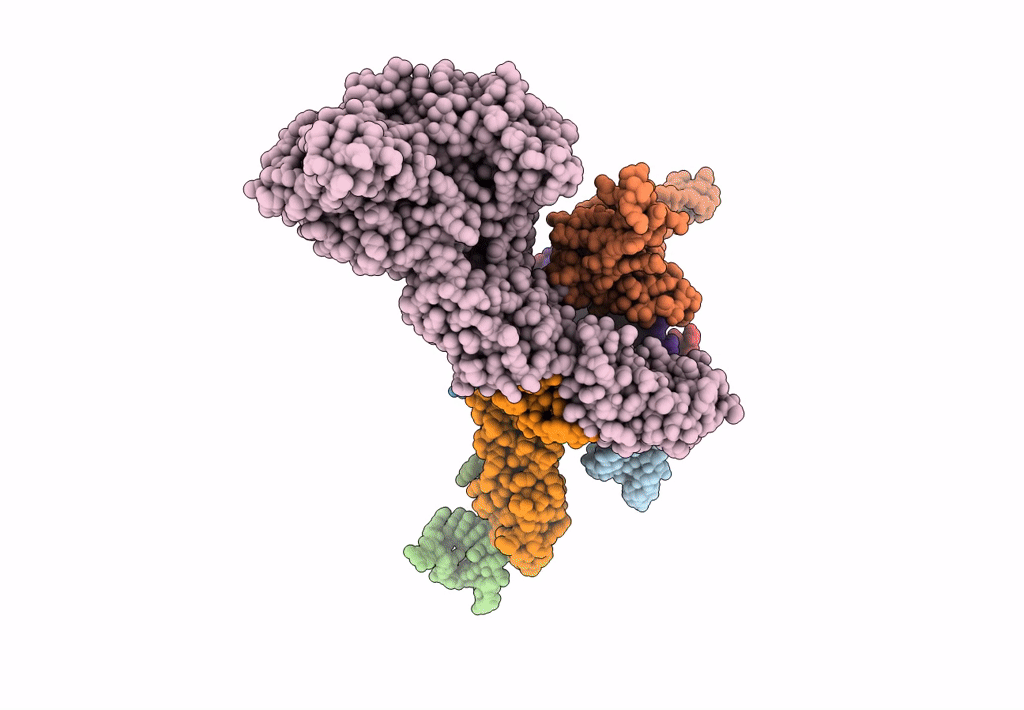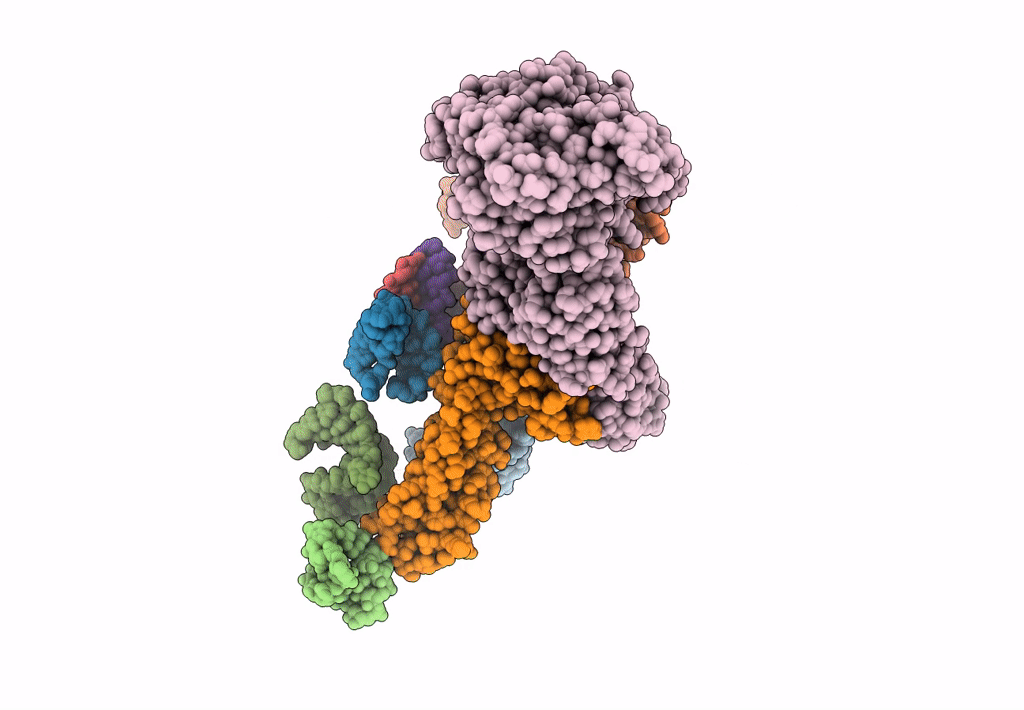
Models for the T. thermophilus ribosome recycling factor and the E. coli elongation factor G bound to the E. coli post-termination complex
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Thermus thermophilus (Taxon ID: 274)
Thermus thermophilus (Taxon ID: 274)
Method Details:
Experimental Method:
Resolution:
9.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE