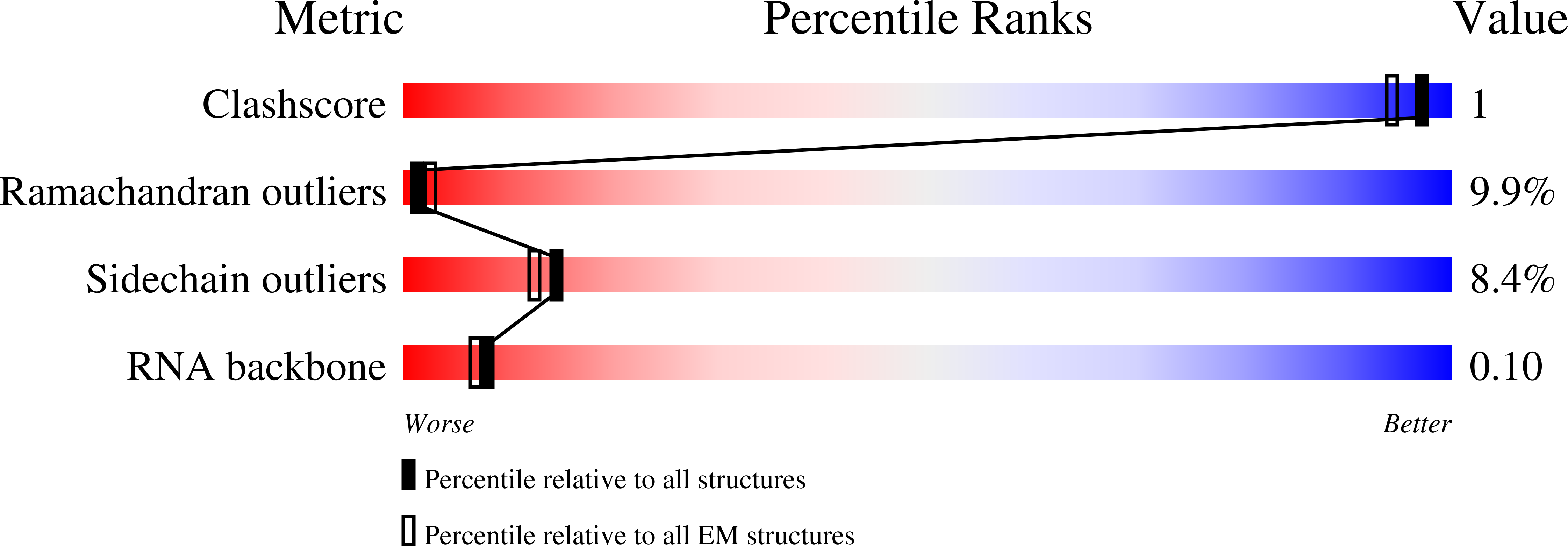
Deposition Date
2010-04-16
Release Date
2010-10-20
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3IYR
Keywords:
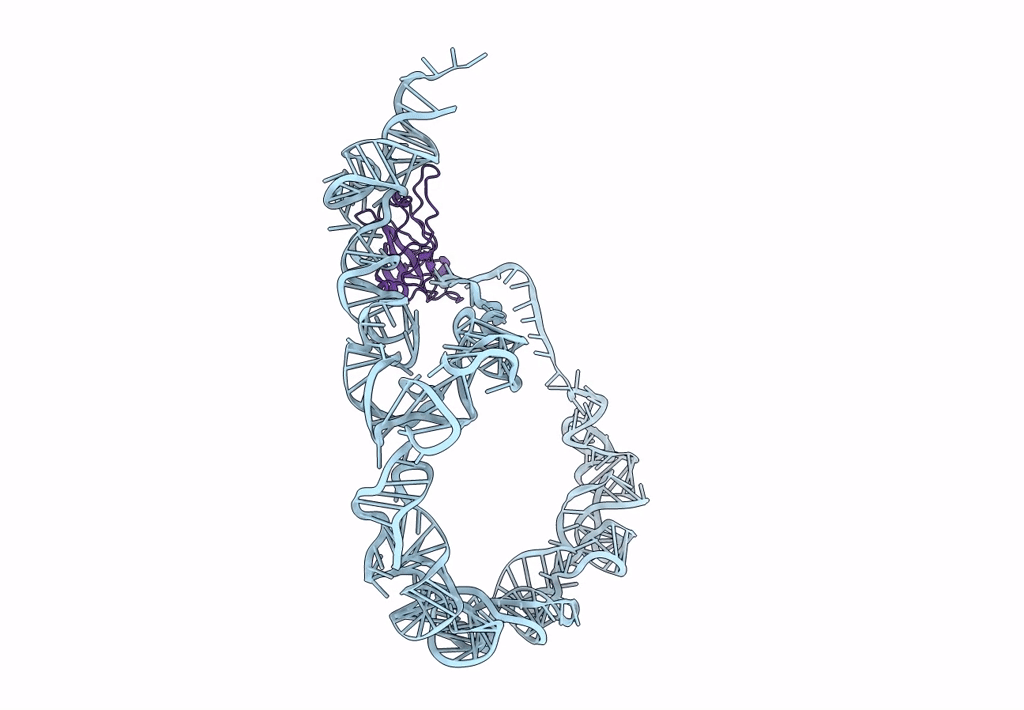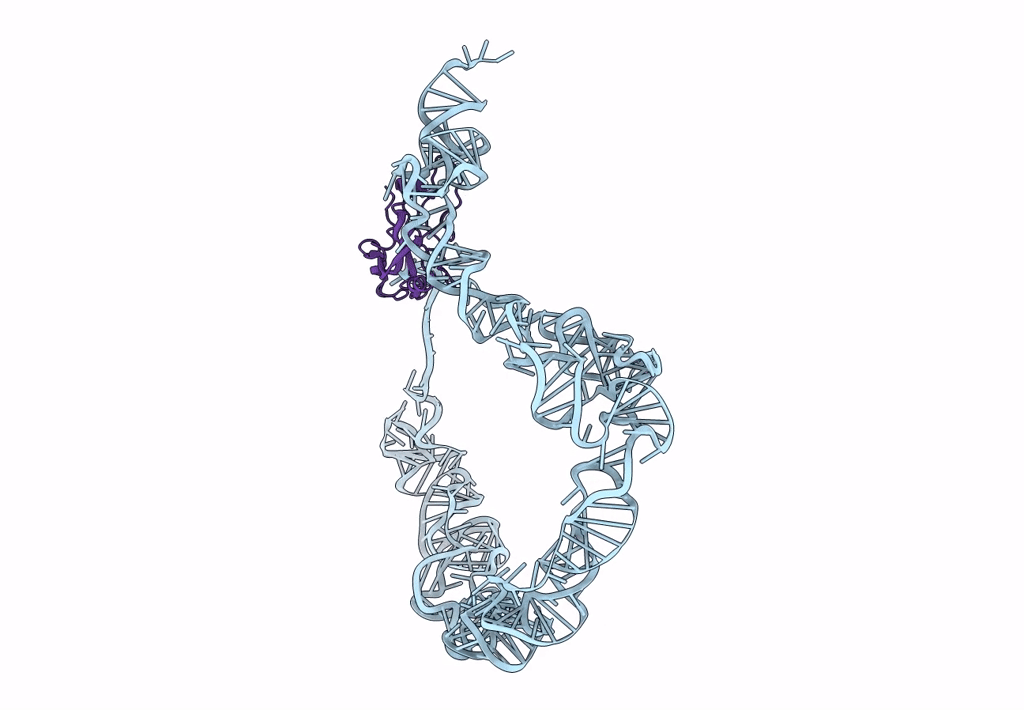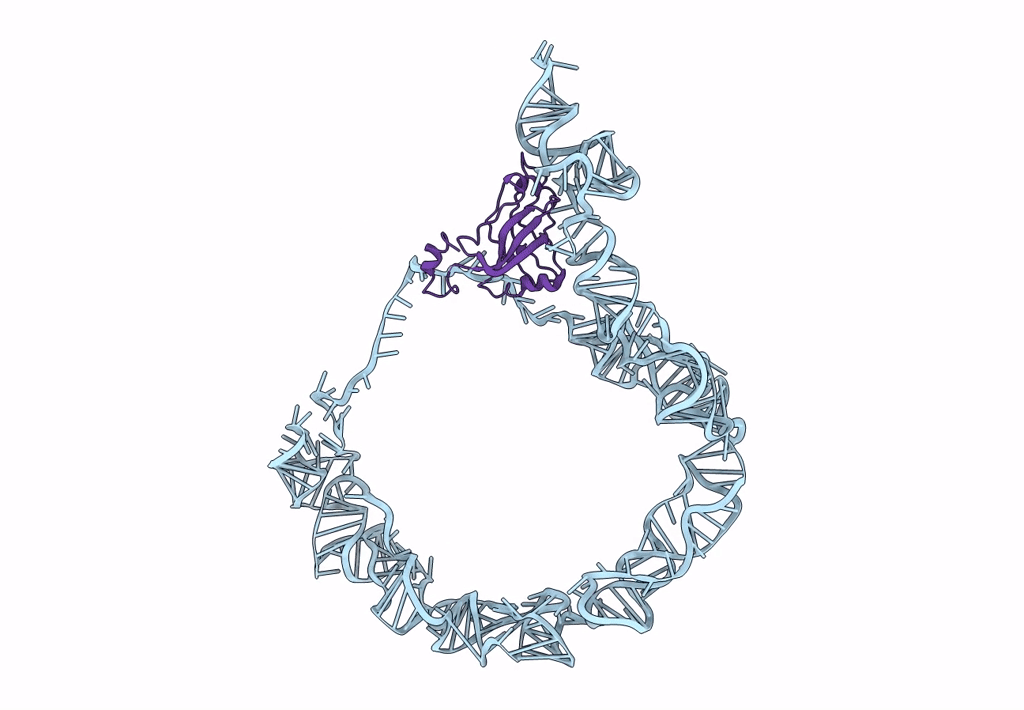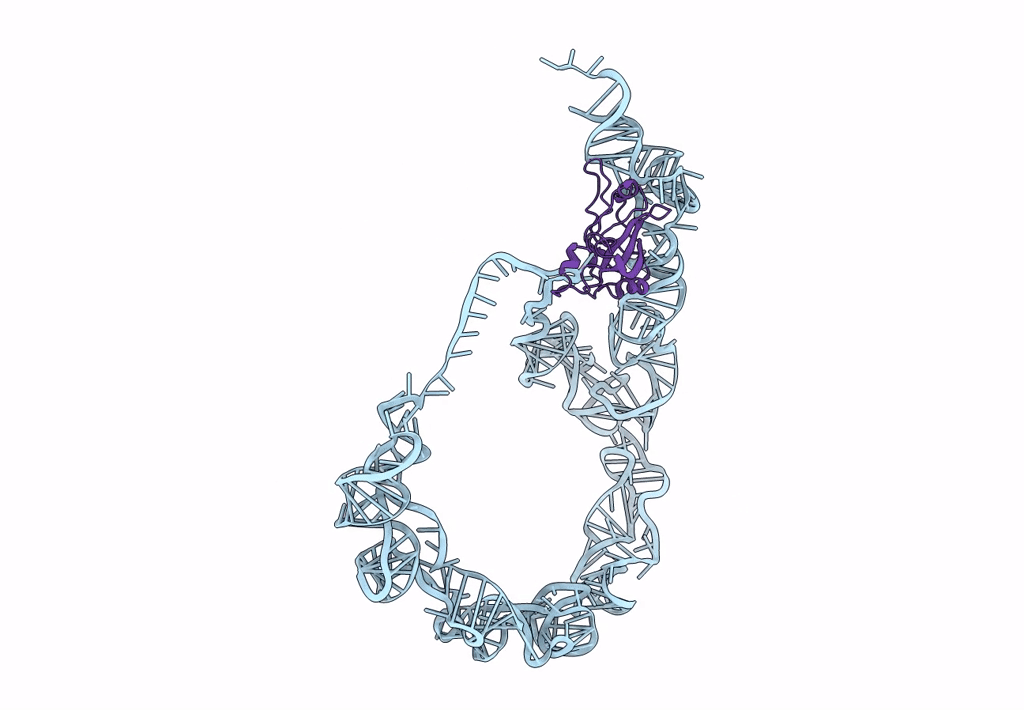
Title:
tmRNA-SmpB: a journey to the center of the bacterial ribosome
Biological Source:
Source Organism(s):
Thermus thermophilus (Taxon ID: 300852)
Method Details:
Experimental Method:
Resolution:
13.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE