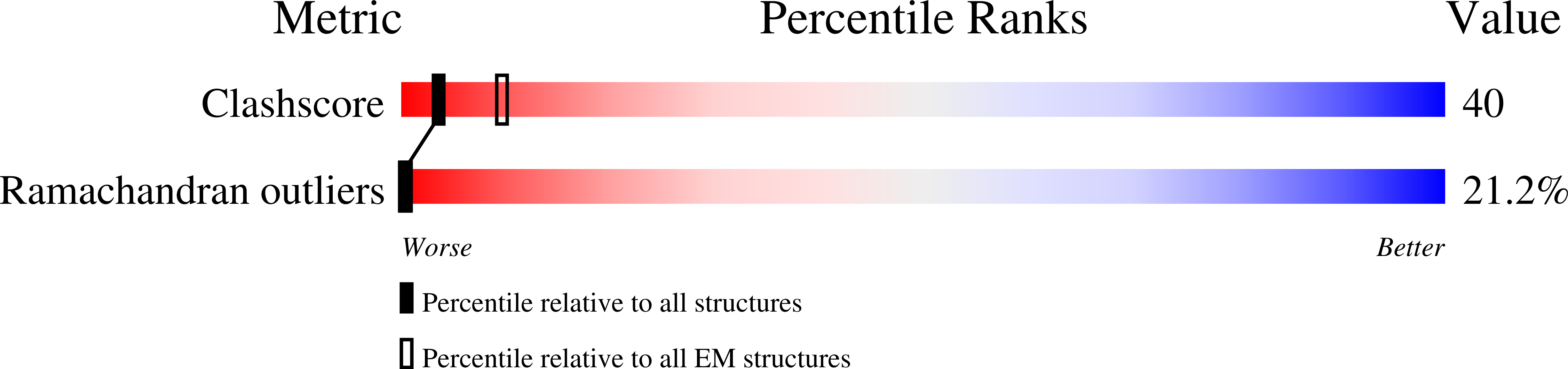
Deposition Date
2010-02-05
Release Date
2010-07-28
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3IYM
Keywords:
Title:
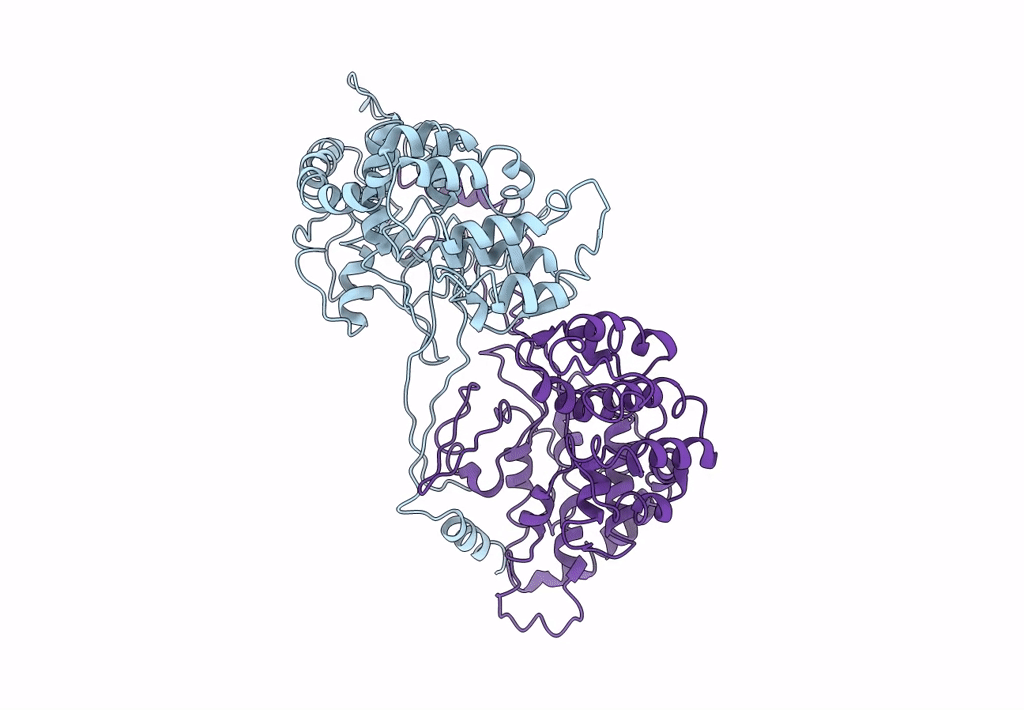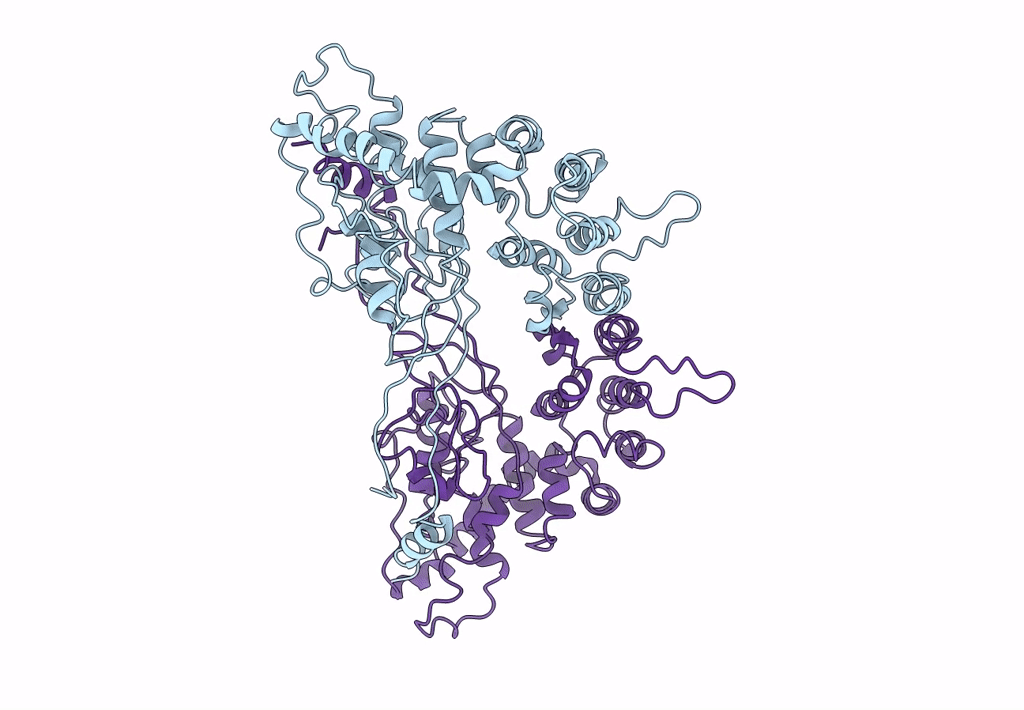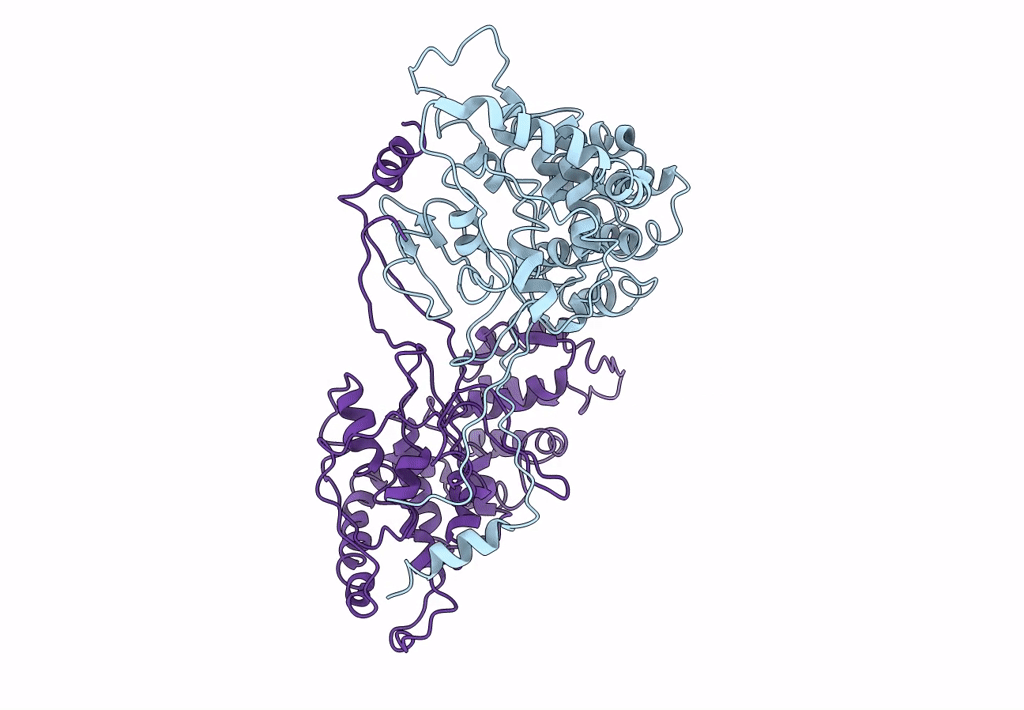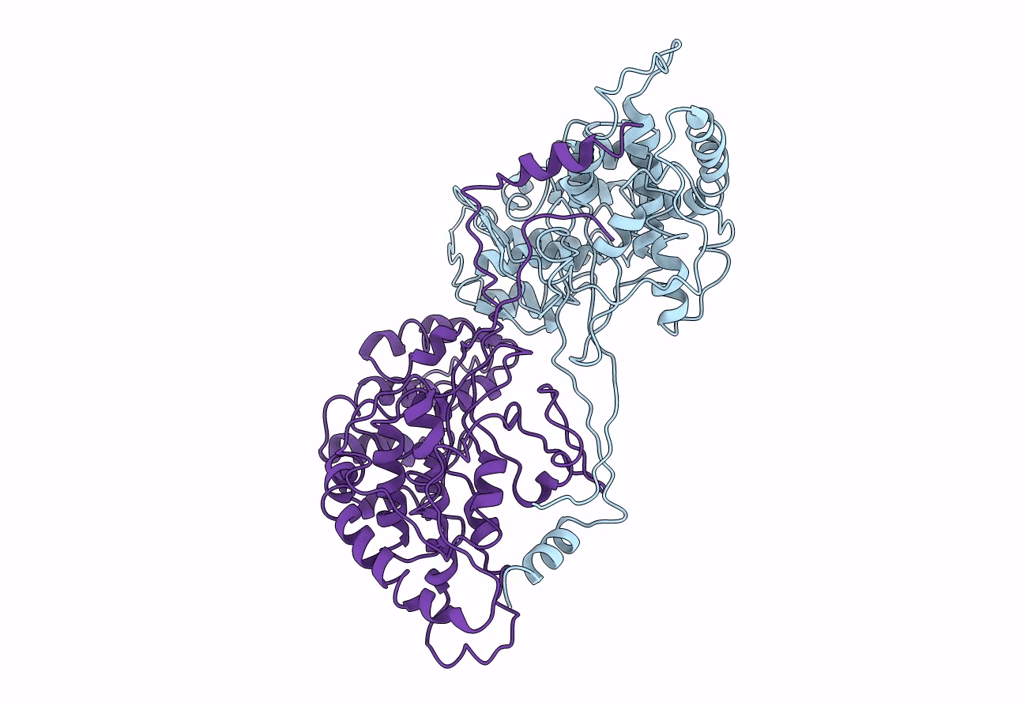
Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling
Biological Source:
Source Organism(s):
Penicillium stoloniferum virus S (Taxon ID: 216371)
Method Details:
Experimental Method:
Resolution:
4.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE