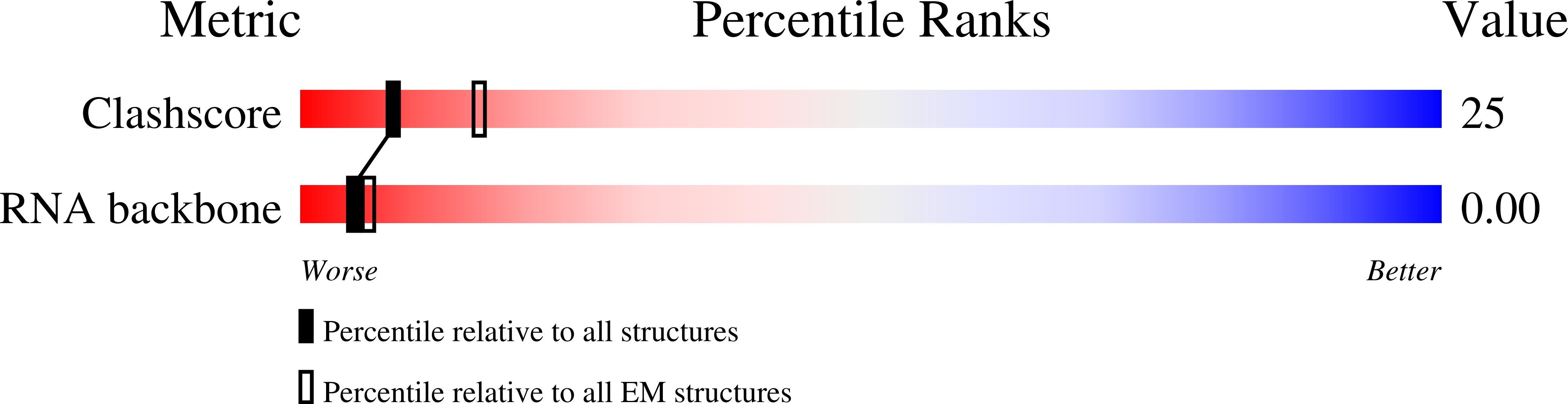Deposition Date
2009-04-20
Release Date
2009-07-07
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3IY9
Keywords:
Title:
Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model
Biological Source:
Source Organism(s):
Leishmania Tarentolae (Taxon ID: 5689)
Homo sapiens (Taxon ID: 9606)
Escherichia coli (Taxon ID: 83333)
Homo sapiens (Taxon ID: 9606)
Escherichia coli (Taxon ID: 83333)
Method Details:
Experimental Method:
Resolution:
14.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE